This notebook is adapted from the NA CORDEX notebook on AWS
http://
ncar -aws -www .s3 -website -us -west -2 .amazonaws .com /plot -zarr -diagnostics .html
Input Data Access¶
This notebook illustrates how to make diagnostic plots using the NA-CORDEX dataset hosted on NCAR’s Geoscience Data Exchange (GDEX)
This data is open access and can be accessed via 2 protocols
HTTPS (if you have access to NCAR’s HPC)
OSDF using an intake-ESM catalog.
# Imports
import intake
import numpy as np
import pandas as pd
import xarray as xr
import seaborn as sns
import re
import matplotlib.pyplot as plt
import osimport fsspec.implementations.http as fshttp
from pelicanfs.core import PelicanFileSystem, PelicanMap, OSDFFileSystem import dask
from dask_jobqueue import PBSCluster
from dask.distributed import Client
from dask.distributed import performance_reportinit_year0 = '1991'
init_year1 = '2020'
final_year0 = '2071'
final_year1 = '2100'# Set up your scratch folder path
username = os.environ["USER"]
glade_scratch = "/glade/derecho/scratch/" + username
print(glade_scratch)/glade/derecho/scratch/harshah
catalog_url = 'https://osdata.gdex.ucar.edu/d316010/catalogs/d316010-osdf-zarr.json'
# catalog_url = '' #NCAR's Object store
print(catalog_url)https://osdata.gdex.ucar.edu/d316010/catalogs/d316010-osdf-zarr.json
Create a PBS cluster¶
# Create a PBS cluster object
cluster = PBSCluster(
job_name = 'dask-wk24-hpc',
cores = 1,
memory = '8GiB',
processes = 1,
local_directory = glade_scratch+'/dask/spill',
log_directory = glade_scratch + '/dask/logs/',
resource_spec = 'select=1:ncpus=1:mem=8GB',
queue = 'casper',
walltime = '5:00:00',
#interface = 'ib0'
interface = 'ext'
)# Create the client to load the Dashboard
client = Client(cluster)
n_workers = 8
cluster.scale(n_workers)
client.wait_for_workers(n_workers = n_workers)
clusterLoading...
Load NA CORDEX data from GDEX using an intake catalog¶
col = intake.open_esm_datastore(catalog_url)
colLoading...
# # Produce a catalog content summary.
# import pprint
# uniques = col.unique(
# columns=["variable", "scenario", "grid", "na-cordex-models", "bias_correction"]
# )
# pprint.pprint(uniques, compact=True, indent=4)Load data into xarray¶
data_var = 'tmax'
col_subset = col.search(
variable=data_var,
grid="NAM-44i",
scenario="eval",
bias_correction="raw",
)
col_subsetLoading...
col_subset.df['path'].valuesarray(['https://osdf-data.gdex.ucar.edu/ncar-gdex/d316010/day/tmax.eval.day.NAM-44i.raw.zarr'],
dtype=object)col_subset['tmax.day.eval.NAM-44i.raw'].dfLoading...
# Load catalog entries for subset into a dictionary of xarray datasets, and open the first one.
dsets = col_subset.to_dataset_dict(zarr_kwargs={"consolidated": True})
print(f"\nDataset dictionary keys:\n {dsets.keys()}")
--> The keys in the returned dictionary of datasets are constructed as follows:
'variable.frequency.scenario.grid.bias_correction'
Loading...
Loading...
Dataset dictionary keys:
dict_keys(['tmax.day.eval.NAM-44i.raw'])
# Load the first dataset and display a summary.
dataset_key = list(dsets.keys())[0]
store_name = dataset_key + ".zarr"
ds = dsets[dataset_key]
dsLoading...
Functions for Plotting¶
Helper Function to Create a Single Map Plot¶
def plotMap(ax, map_slice, date_object=None, member_id=None):
"""Create a map plot on the given axes, with min/max as text"""
ax.imshow(map_slice, origin='lower')
minval = map_slice.min(dim = ['lat', 'lon'])
maxval = map_slice.max(dim = ['lat', 'lon'])
# Format values to have at least 4 digits of precision.
ax.text(0.01, 0.03, "Min: %3g" % minval, transform=ax.transAxes, fontsize=12)
ax.text(0.99, 0.03, "Max: %3g" % maxval, transform=ax.transAxes, fontsize=12, horizontalalignment='right')
ax.set_xticks([])
ax.set_yticks([])
if date_object:
ax.set_title(date_object.values.astype(str)[:10], fontsize=12)
if member_id:
ax.set_ylabel(member_id, fontsize=12)
return axHelper Function for Finding Dates with Available Data¶
def getValidDateIndexes(member_slice):
"""Search for the first and last dates with finite values."""
min_values = member_slice.min(dim = ['lat', 'lon'])
is_finite = np.isfinite(min_values)
finite_indexes = np.where(is_finite)
start_index = finite_indexes[0][0]
end_index = finite_indexes[0][-1]
return start_index, end_indexFunction Producing Maps of First, Middle, and Final Timesteps¶
def plot_first_mid_last(ds, data_var, store_name):
"""Plot the first, middle, and final time steps for several climate runs."""
num_members_to_plot = 4
member_names = ds.coords['member_id'].values[0:num_members_to_plot]
figWidth = 18
figHeight = 12
numPlotColumns = 3
fig, axs = plt.subplots(num_members_to_plot, numPlotColumns, figsize=(figWidth, figHeight), constrained_layout=True)
for index in np.arange(num_members_to_plot):
mem_id = member_names[index]
data_slice = ds[data_var].sel(member_id=mem_id)
start_index, end_index = getValidDateIndexes(data_slice)
midDateIndex = np.floor(len(ds.time) / 2).astype(int)
startDate = ds.time[start_index]
first_step = data_slice.sel(time=startDate)
ax = axs[index, 0]
plotMap(ax, first_step, startDate, mem_id)
midDate = ds.time[midDateIndex]
mid_step = data_slice.sel(time=midDate)
ax = axs[index, 1]
plotMap(ax, mid_step, midDate)
endDate = ds.time[end_index]
last_step = data_slice.sel(time=endDate)
ax = axs[index, 2]
plotMap(ax, last_step, endDate)
plt.suptitle(f'First, Middle, and Last Timesteps for Selected Runs in "{store_name}"', fontsize=20)
return figFunction Producing Statistical Map Plots¶
def plot_stat_maps(ds, data_var, store_name):
"""Plot the mean, min, max, and standard deviation values for several climate runs, aggregated over time."""
num_members_to_plot = 4
member_names = ds.coords['member_id'].values[0:num_members_to_plot]
figWidth = 25
figHeight = 12
numPlotColumns = 4
fig, axs = plt.subplots(num_members_to_plot, numPlotColumns, figsize=(figWidth, figHeight), constrained_layout=True)
for index in np.arange(num_members_to_plot):
mem_id = member_names[index]
data_slice = ds[data_var].sel(member_id=mem_id)
data_agg = data_slice.min(dim='time')
plotMap(axs[index, 0], data_agg, member_id=mem_id)
data_agg = data_slice.max(dim='time')
plotMap(axs[index, 1], data_agg)
data_agg = data_slice.mean(dim='time')
plotMap(axs[index, 2], data_agg)
data_agg = data_slice.std(dim='time')
plotMap(axs[index, 3], data_agg)
axs[0, 0].set_title(f'min({data_var})', fontsize=15)
axs[0, 1].set_title(f'max({data_var})', fontsize=15)
axs[0, 2].set_title(f'mean({data_var})', fontsize=15)
axs[0, 3].set_title(f'std({data_var})', fontsize=15)
plt.suptitle(f'Spatial Statistics for Selected Runs in "{store_name}"', fontsize=20)
return figFunction Producing Time Series Plots¶
Also show which dates have no available data values, as a rug plot.
def plot_timeseries(ds, data_var, store_name):
"""Plot the mean, min, max, and standard deviation values for several climate runs,
aggregated over lat/lon dimensions."""
num_members_to_plot = 4
member_names = ds.coords['member_id'].values[0:num_members_to_plot]
figWidth = 25
figHeight = 20
linewidth = 0.5
numPlotColumns = 1
fig, axs = plt.subplots(num_members_to_plot, numPlotColumns, figsize=(figWidth, figHeight))
for index in np.arange(num_members_to_plot):
mem_id = member_names[index]
data_slice = ds[data_var].sel(member_id=mem_id)
unit_string = ds[data_var].attrs['units']
min_vals = data_slice.min(dim = ['lat', 'lon'])
max_vals = data_slice.max(dim = ['lat', 'lon'])
mean_vals = data_slice.mean(dim = ['lat', 'lon'])
std_vals = data_slice.std(dim = ['lat', 'lon'])
missing_indexes = np.isnan(min_vals)
missing_times = ds.time[missing_indexes]
axs[index].plot(ds.time, max_vals, linewidth=linewidth, label='max', color='red')
axs[index].plot(ds.time, mean_vals, linewidth=linewidth, label='mean', color='black')
axs[index].fill_between(ds.time, (mean_vals - std_vals), (mean_vals + std_vals),
color='grey', linewidth=0, label='std', alpha=0.5)
axs[index].plot(ds.time, min_vals, linewidth=linewidth, label='min', color='blue')
ymin, ymax = axs[index].get_ylim()
rug_y = ymin + 0.01*(ymax-ymin)
axs[index].plot(missing_times, [rug_y]*len(missing_times), '|', color='m', label='missing')
axs[index].set_title(mem_id, fontsize=20)
axs[index].legend(loc='upper right')
axs[index].set_ylabel(unit_string)
plt.tight_layout(pad=10.2, w_pad=3.5, h_pad=3.5)
plt.suptitle(f'Temporal Statistics for Selected Runs in "{store_name}"', fontsize=20)
return figProduce Diagnostic Plots¶
Plot First, Middle, and Final Timesteps for Several Output Runs (less compute intensive)¶
%%time
# Plot using the Zarr Store obtained from an earlier step in the notebook.
figure = plot_first_mid_last(ds, data_var, store_name)
plt.show()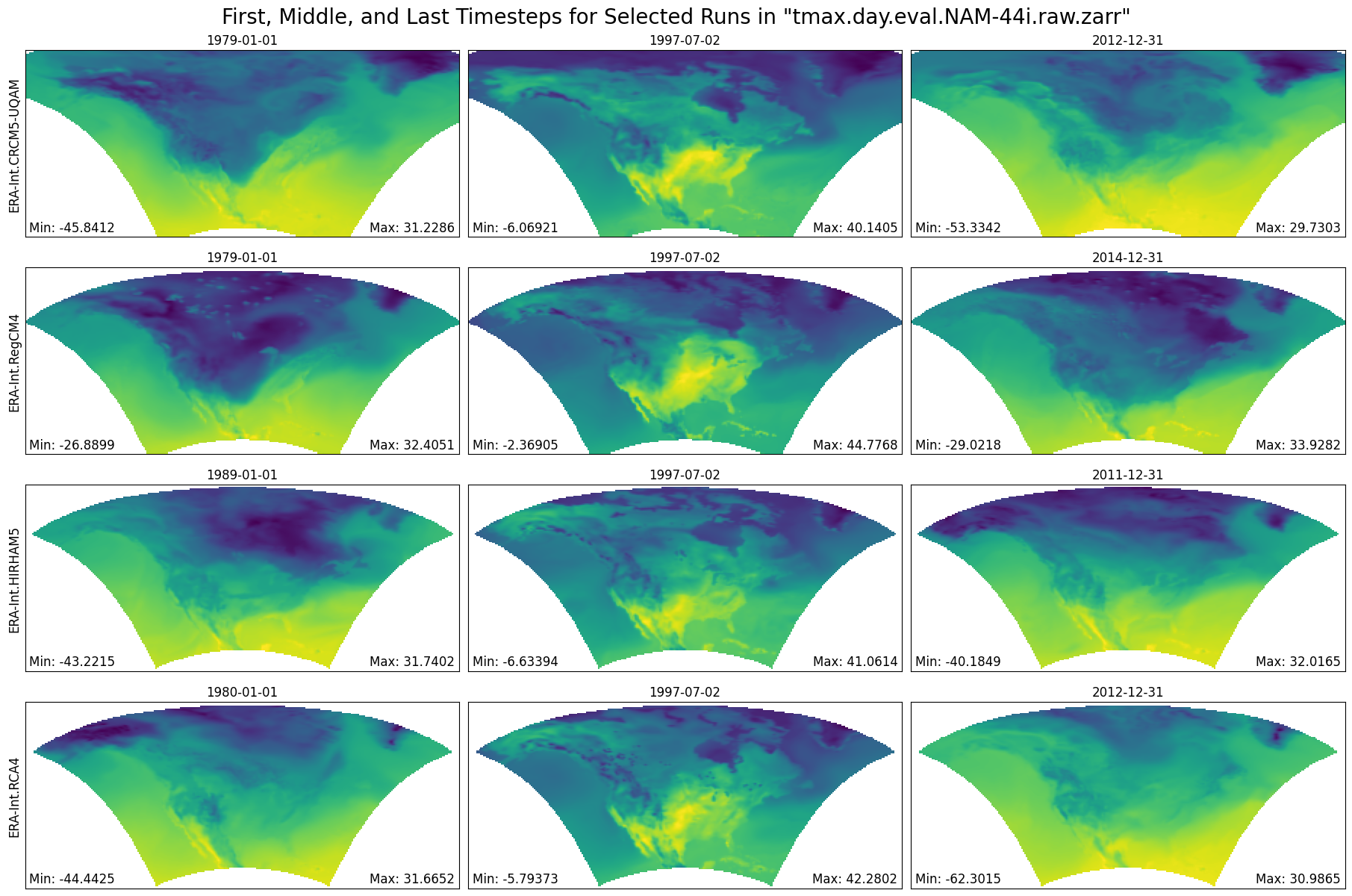
CPU times: user 4.96 s, sys: 419 ms, total: 5.38 s
Wall time: 43.9 s
cluster.close()