
Dask DataFrame#
ESDS dask tutorial | 06 February, 2023
Negin Sobhani, Brian Vanderwende, Deepak Cherian, Ben Kirk
Computational & Information Systems Lab (CISL)
negins@ucar.edu, vanderwb@ucar.edu
In this tutorial, you learn:#
Basic concepts and features of Dask DataFrames
Applications of Dask DataFrames
Interacting with Dask DataFrames
Built-in operations with Dask DataFrames
Dask DataFrames Best Practices
Introduction#
pandas is a very popular tool for working with tabular datasets, but the dataset needs to fit into the memory.
pandas operates best with smaller datasets, and if you have a large dataset, you’ll receive an out of memory error using pandas. A general rule of thumb for pandas is:
“Have 5 to 10 times as much RAM as the size of your dataset”
~ Wes McKinney (2017) in 10 things I hate about pandas
But Dask DataFrame can be used to solve pandas performance issues with larger-than-memory datasets.
What is Dask DataFrame?#
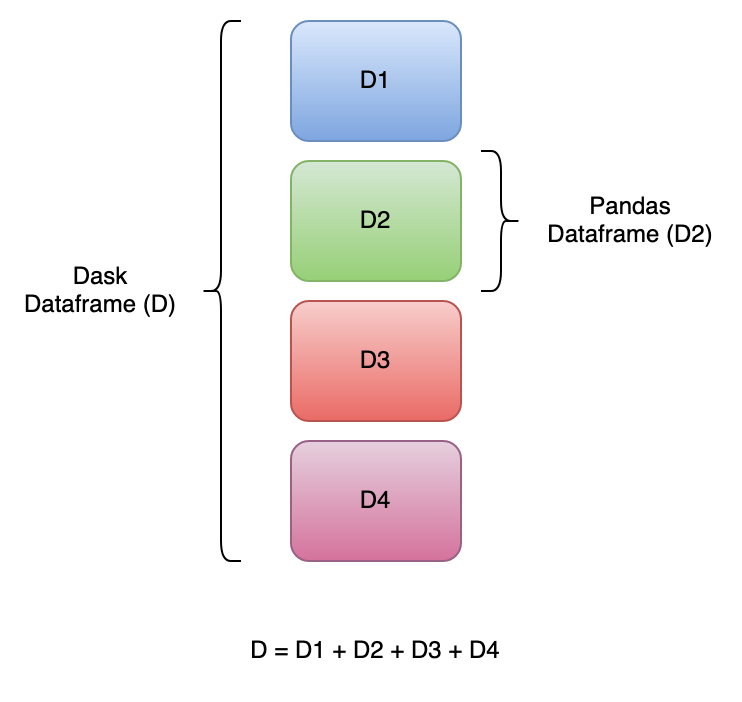
A Dask DataFrame is a parallel DataFrame composed of smaller pandas DataFrames (also known as partitions).
Dask Dataframes look and feel like the pandas DataFrames on the surface.
Dask DataFrames partition the data into manageable partitions that can be processed in parallel and across multiple cores or computers.
Similar to Dask Arrays, Dask DataFrames are lazy!
Unlike pandas, operations on Dask DataFrames are not computed until you explicitly request them (e.g. by calling
.compute).
When to use Dask DataFrame and when to avoid it?#
Dask DataFrames are used in situations where pandas fails or has poor performance due to data size.
Dask DataFrame is a good choice when doing parallalizeable computations.
Some examples are:
Element-wise operations such as
df.x + df.yRow-wise filtering such as
df[df.x>0]Common aggregations such as
df.x.max()Dropping duplicates such as
df.x.drop_duplicate()
However, Dask is not great for operations that requires shuffling or re-indexing.
Some examples are:
Set index:
df.set_index(df.x)
See the Dask DataFrame API documentation for a compehnsive list of available functions.
Tutorial Dataset#
In this tutorial, we are going to use the NOAA Global Historical Climatology Network Daily (GHCN-D) dataset.
GHCN-D is a public available dataset that includes daily climate records from +100,000 surface observations around the world.
This is an example of a real dataset that is used by NCAR scientists for their research. GHCN-D raw dataset for all stations is available through NOAA Climate Data Online.
To learn more about GHCNd dataset, please visit:
Download the data#
For this example, we are going to look through a subset of data from the GHCN-D dataset.
First, we look at the daily observations from Denver International Airport, next we are going to look through selected stations in the US.
The access the preprocessed dataset for this tutorial, please run the following script:
!./get_data.sh
--2023-02-05 13:50:53-- https://docs.google.com/uc?export=download&id=14doSRn8hT14QYtjZz28GKv14JgdIsbFF
Resolving docs.google.com... 142.250.72.78, 2607:f8b0:400f:807::200e
Connecting to docs.google.com|142.250.72.78|:443... connected.
HTTP request sent, awaiting response... 303 See Other
Location: https://doc-08-74-docs.googleusercontent.com/docs/securesc/ha0ro937gcuc7l7deffksulhg5h7mbp1/hkbhr7uhov17jj84626eu6oouerldgvu/1675630200000/12706685253353286515/*/14doSRn8hT14QYtjZz28GKv14JgdIsbFF?e=download&uuid=8c5eb41b-5044-4497-8562-d31dc9d3bc4d [following]
Warning: wildcards not supported in HTTP.
--2023-02-05 13:51:05-- https://doc-08-74-docs.googleusercontent.com/docs/securesc/ha0ro937gcuc7l7deffksulhg5h7mbp1/hkbhr7uhov17jj84626eu6oouerldgvu/1675630200000/12706685253353286515/*/14doSRn8hT14QYtjZz28GKv14JgdIsbFF?e=download&uuid=8c5eb41b-5044-4497-8562-d31dc9d3bc4d
Resolving doc-08-74-docs.googleusercontent.com... 142.250.72.33, 2607:f8b0:400f:804::2001
Connecting to doc-08-74-docs.googleusercontent.com|142.250.72.33|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 19013376 (18M) [application/x-tar]
Saving to: ‘../data.tar.gz’
../data.tar.gz 100%[===================>] 18.13M 52.5MB/s in 0.3s
2023-02-05 13:51:05 (52.5 MB/s) - ‘../data.tar.gz’ saved [19013376/19013376]
USC00023160.csv
USC00027281.csv
USC00027390.csv
USC00030936.csv
USC00031596.csv
USC00032444.csv
USC00035186.csv
USC00035754.csv
USC00035820.csv
USC00035908.csv
USC00042294.csv
USC00044259.csv
USC00048758.csv
USC00050848.csv
USC00051294.csv
USC00051528.csv
USC00051564.csv
USC00051741.csv
USC00052184.csv
USC00052281.csv
USC00052446.csv
USC00053005.csv
USC00053038.csv
USC00053146.csv
USC00053662.csv
USC00053951.csv
USC00054076.csv
USC00054770.csv
USC00054834.csv
USC00055322.csv
USC00055722.csv
USC00057167.csv
USC00057337.csv
--2023-02-05 13:51:07-- https://docs.google.com/uc?export=download&id=15rCwQUxxpH6angDhpXzlvbe1nGetYHrf
Resolving docs.google.com... 142.250.72.78, 2607:f8b0:400f:807::200e
Connecting to docs.google.com|142.250.72.78|:443... connected.
HTTP request sent, awaiting response... 303 See Other
Location: https://doc-0o-74-docs.googleusercontent.com/docs/securesc/ha0ro937gcuc7l7deffksulhg5h7mbp1/9f30ek7ivt0gum0c34hgalhsikads9th/1675630275000/12706685253353286515/*/15rCwQUxxpH6angDhpXzlvbe1nGetYHrf?e=download&uuid=deb6d161-3a18-444b-b54a-a6ee1280cba9 [following]
Warning: wildcards not supported in HTTP.
--2023-02-05 13:51:15-- https://doc-0o-74-docs.googleusercontent.com/docs/securesc/ha0ro937gcuc7l7deffksulhg5h7mbp1/9f30ek7ivt0gum0c34hgalhsikads9th/1675630275000/12706685253353286515/*/15rCwQUxxpH6angDhpXzlvbe1nGetYHrf?e=download&uuid=deb6d161-3a18-444b-b54a-a6ee1280cba9
Resolving doc-0o-74-docs.googleusercontent.com... 142.250.72.33, 2607:f8b0:400f:804::2001
Connecting to doc-0o-74-docs.googleusercontent.com|142.250.72.33|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 20132755 (19M) [application/x-tar]
Saving to: ‘../data.tar.gz’
../data.tar.gz 100%[===================>] 19.20M 67.9MB/s in 0.3s
2023-02-05 13:51:16 (67.9 MB/s) - ‘../data.tar.gz’ saved [20132755/20132755]
USC00057936.csv
USC00058204.csv
USC00058429.csv
USC00059243.csv
USC00068138.csv
USC00080211.csv
USC00084731.csv
USC00088824.csv
USC00098703.csv
USC00100010.csv
USC00100470.csv
USC00105275.csv
USC00106152.csv
USC00107264.csv
USC00108137.csv
USC00110338.csv
USC00112140.csv
USC00112193.csv
USC00112348.csv
USC00112483.csv
USC00113335.csv
USC00114108.csv
USC00114442.csv
USC00114823.csv
USC00115079.csv
USC00115326.csv
USC00115712.csv
USC00115768.csv
USC00115833.csv
USC00115901.csv
USC00115943.csv
USC00116446.csv
USW00003017.csv
This script should save the preprocessed GHCN-D data in ../data path.
Pandas DataFrame Basics#
Let’s start with an example using pandas DataFrame.
First, let’s read in the comma-seperated GHCN-D dataset for one station at Denver International Airport (DIA), CO (site ID : USW00003017).
To see the list of all available GHCN-D sites and their coordinates and IDs, please see this link.
import os
import pandas as pd
# DIA ghcnd id
site = 'USW00003017'
data_dir = '../data/'
df = pd.read_csv(os.path.join(data_dir, site+'.csv'), parse_dates=['DATE'], index_col=0)
# Display the top five rows of the dataframe
df.head()
| ID | YEAR | MONTH | DAY | TMAX | TMAX_FLAGS | TMIN | TMIN_FLAGS | PRCP | PRCP_FLAGS | ... | RHMN_FLAGS | RHMX | RHMX_FLAGS | PSUN | PSUN_FLAGS | LATITUDE | LONGITUDE | ELEVATION | STATE | STATION | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DATE | |||||||||||||||||||||
| 1994-07-20 | USW00003017 | 1994 | 7 | 20 | 316.0 | XXS | 150.0 | XXS | 20.0 | DXS | ... | XXX | NaN | XXX | NaN | XXX | 39.8467 | -104.6561 | 1647.1 | CO | DENVER INTL AP |
| 1994-07-23 | USW00003017 | 1994 | 7 | 23 | 355.0 | XXS | 166.0 | XXS | 0.0 | DXS | ... | XXX | NaN | XXX | NaN | XXX | 39.8467 | -104.6561 | 1647.1 | CO | DENVER INTL AP |
| 1994-07-24 | USW00003017 | 1994 | 7 | 24 | 333.0 | XXS | 155.0 | XXS | 81.0 | DXS | ... | XXX | NaN | XXX | NaN | XXX | 39.8467 | -104.6561 | 1647.1 | CO | DENVER INTL AP |
| 1994-07-25 | USW00003017 | 1994 | 7 | 25 | 327.0 | XXS | 172.0 | XXS | 0.0 | DXS | ... | XXX | NaN | XXX | NaN | XXX | 39.8467 | -104.6561 | 1647.1 | CO | DENVER INTL AP |
| 1994-07-26 | USW00003017 | 1994 | 7 | 26 | 327.0 | XXS | 155.0 | XXS | 0.0 | DXS | ... | XXX | NaN | XXX | NaN | XXX | 39.8467 | -104.6561 | 1647.1 | CO | DENVER INTL AP |
5 rows × 99 columns
Question: What variables are available?
df.columns
Index(['ID', 'YEAR', 'MONTH', 'DAY', 'TMAX', 'TMAX_FLAGS', 'TMIN',
'TMIN_FLAGS', 'PRCP', 'PRCP_FLAGS', 'TAVG', 'TAVG_FLAGS', 'SNOW',
'SNOW_FLAGS', 'SNWD', 'SNWD_FLAGS', 'AWND', 'AWND_FLAGS', 'FMTM',
'FMTM_FLAGS', 'PGTM', 'PGTM_FLAGS', 'WDF2', 'WDF2_FLAGS', 'WDF5',
'WDF5_FLAGS', 'WSF2', 'WSF2_FLAGS', 'WSF5', 'WSF5_FLAGS', 'WT01',
'WT01_FLAGS', 'WT02', 'WT02_FLAGS', 'WT08', 'WT08_FLAGS', 'WT16',
'WT16_FLAGS', 'WT17', 'WT17_FLAGS', 'WT18', 'WT18_FLAGS', 'WT03',
'WT03_FLAGS', 'WT05', 'WT05_FLAGS', 'WT19', 'WT19_FLAGS', 'WT10',
'WT10_FLAGS', 'WT09', 'WT09_FLAGS', 'WT06', 'WT06_FLAGS', 'WT07',
'WT07_FLAGS', 'WT11', 'WT11_FLAGS', 'WT13', 'WT13_FLAGS', 'WT21',
'WT21_FLAGS', 'WT14', 'WT14_FLAGS', 'WT15', 'WT15_FLAGS', 'WT22',
'WT22_FLAGS', 'WT04', 'WT04_FLAGS', 'WV03', 'WV03_FLAGS', 'TSUN',
'TSUN_FLAGS', 'WV01', 'WV01_FLAGS', 'WESD', 'WESD_FLAGS', 'ADPT',
'ADPT_FLAGS', 'ASLP', 'ASLP_FLAGS', 'ASTP', 'ASTP_FLAGS', 'AWBT',
'AWBT_FLAGS', 'RHAV', 'RHAV_FLAGS', 'RHMN', 'RHMN_FLAGS', 'RHMX',
'RHMX_FLAGS', 'PSUN', 'PSUN_FLAGS', 'LATITUDE', 'LONGITUDE',
'ELEVATION', 'STATE', 'STATION'],
dtype='object')
The description and units of the dataset is available here.
Operations on pandas DataFrame#
pandas DataFrames has several features that give us flexibility to do different calculations and analysis on our dataset. Let’s check some out:
Simple Analysis#
For example:
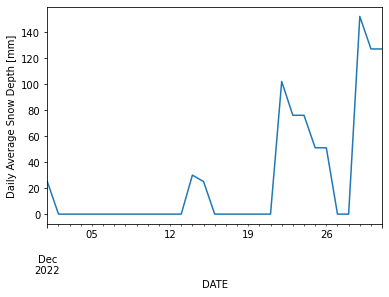
When was the coldest day at this station during December of last year?
# use python slicing notation inside .loc
# use idxmin() to find the index of minimum valus
df.loc['2022-12-01':'2022-12-31'].TMIN.idxmin()
Timestamp('2022-12-22 00:00:00')
# Here we easily plot the prior data using matplotlib from pandas
# -- .loc for value based indexing
df.loc['2022-12-01':'2022-12-31'].SNWD.plot(ylabel= 'Daily Average Snow Depth [mm]')
<AxesSubplot:xlabel='DATE', ylabel='Daily Average Snow Depth [mm]'>
How many snow days do we have each year at this station?
Pandas groupby is used for grouping the data according to the categories.
# 1- First select days with snow > 0
# 2- Create a "groupby object" based on the selected columns
# 3- use .size() to compute the size of each group
# 4- sort the values descending
# we count days where SNOW>0, and sort them and show top 5 years:
df[df['SNOW']>0].groupby('YEAR').size().sort_values(ascending=False).head()
YEAR
2015 36
2019 34
2014 32
2008 32
2007 31
dtype: int64
Or for a more complex analysis:
For example, we have heard that this could be Denver’s first January in 13 years with no 60-degree days.
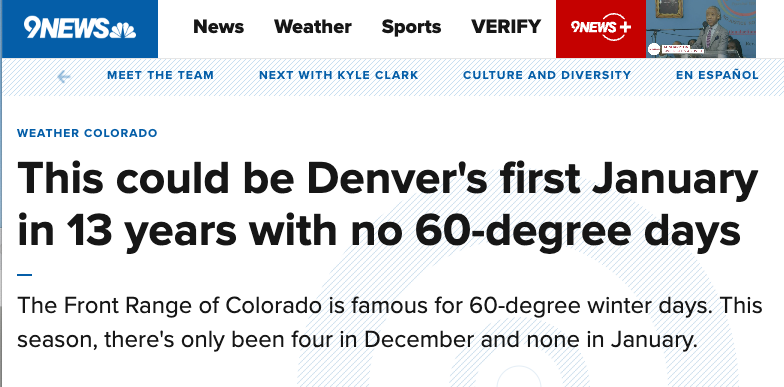
Below, we show all days with high temperature above 60°F (155.5°C/10) since 2010:
df[(df['MONTH']==1) & (df['YEAR']>=2010) & (df['TMAX']>155.5)].groupby(['YEAR']).size()
YEAR
2011 1
2012 6
2013 4
2014 3
2015 6
2016 1
2017 4
2018 5
2019 3
2020 2
2021 2
2022 3
dtype: int64
This is great! But how big is this dataset for one station?
First, let’s check the file size:
!ls -lh ../data/USW00003017.csv
-rw-r--r-- 1 negins ncar 3.6M Feb 5 07:14 ../data/USW00003017.csv
Similar to the previous tutorial, we can use the following function to find the size of a variable on memory.
# Define function to display variable size in MB
import sys
def var_size(in_var):
result = sys.getsizeof(in_var) / 1e6
print(f"Size of variable: {result:.2f} MB")
var_size(df)
Size of variable: 33.21 MB
Remember, the above rule?
“Have 5 to 10 times as much RAM as the size of your dataset”
~ Wes McKinney (2017) in 10 things I hate about pandas
So far, we read in and analyzed data for one station. We have a total of +118,000 stations over the world and +4500 stations in Colorado alone!
What if we want to look at the larger dataset?
Scaling up to a larger dataset#
Let’s start by reading data from selected stations. The downloaded data for this example includes the climatology observations from 66 selected sites in Colorado.
Pandas can concatenate data to load data spread across multiple files:
!du -csh ../data/*.csv |tail -n1
566M total
Using a for loop with pandas.concat, we can read multiple files at the same time:
%%time
import glob
co_sites = glob.glob(os.path.join(data_dir, '*.csv'))
df = pd.concat(pd.read_csv(f, index_col=0, parse_dates=['DATE']) for f in co_sites)
CPU times: user 11.9 s, sys: 2.87 s, total: 14.8 s
Wall time: 19.5 s
How many stations have we read in?
print ("Concatenated data for", len(df.ID.unique()), "unique sites.")
Concatenated data for 66 unique sites.
Now that we concatenated the data for all sites in one DataFrame, we can do similar analysis on it:
Which site has recorded the most snow days in a year?
%%time
# ~90s on 4GB RAM
snowy_days = df[df['SNOW']>0].groupby(['ID','YEAR']).size()
print ('This site has the highest number of snow days in a year : ')
snowy_days.agg(['idxmax','max'])
This site has the highest number of snow days in a year :
CPU times: user 4.49 s, sys: 15.4 s, total: 19.9 s
Wall time: 1min 21s
idxmax (USC00052281, 1983)
max 102
dtype: object
Excersise: Which Colorado site has recorded the most snow days in 2023?
Dask allows us to conceptualize all of these files as a single dataframe!
# Let's do a little cleanup
del df, snowy_days
Computations on Dask DataFrame#
Create a “LocalCluster” Client with Dask#
from dask.distributed import Client, LocalCluster
cluster = LocalCluster()
client = Client(cluster)
client
Client
Client-53ec3d8d-a599-11ed-bb54-3cecef1b11d4
| Connection method: Cluster object | Cluster type: distributed.LocalCluster |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/negins/proxy/8787/status |
Cluster Info
LocalCluster
2ddb751e
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/negins/proxy/8787/status | Workers: 4 |
| Total threads: 4 | Total memory: 4.00 GiB |
| Status: running | Using processes: True |
Scheduler Info
Scheduler
Scheduler-7ff96b2c-e98a-4ea8-9644-554701393a83
| Comm: tcp://127.0.0.1:38113 | Workers: 4 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/negins/proxy/8787/status | Total threads: 4 |
| Started: Just now | Total memory: 4.00 GiB |
Workers
Worker: 0
| Comm: tcp://127.0.0.1:35345 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/negins/proxy/40352/status | Memory: 1.00 GiB |
| Nanny: tcp://127.0.0.1:44090 | |
| Local directory: /glade/scratch/negins/dask-worker-space/worker-5nmxv_zk | |
Worker: 1
| Comm: tcp://127.0.0.1:39634 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/negins/proxy/46672/status | Memory: 1.00 GiB |
| Nanny: tcp://127.0.0.1:39719 | |
| Local directory: /glade/scratch/negins/dask-worker-space/worker-f5__nvpk | |
Worker: 2
| Comm: tcp://127.0.0.1:37524 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/negins/proxy/33848/status | Memory: 1.00 GiB |
| Nanny: tcp://127.0.0.1:37649 | |
| Local directory: /glade/scratch/negins/dask-worker-space/worker-a517utir | |
Worker: 3
| Comm: tcp://127.0.0.1:46848 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/negins/proxy/36957/status | Memory: 1.00 GiB |
| Nanny: tcp://127.0.0.1:40787 | |
| Local directory: /glade/scratch/negins/dask-worker-space/worker-mboofx4v | |
☝️ Click the Dashboard link above.
👈 Or click the “Search” 🔍 button in the dask-labextension dashboard.
Dask DataFrame read_csv to read multiple files#
dask.dataframe.read_csv function can be used in conjunction with glob to read multiple csv files at the same time.
Remember we can read one file with pandas.read_csv. For reading multiple files with pandas, we have to concatenate them with pd.concatenate. However, we can read many files at once just using dask.dataframe.read_csv.
Overall, Dask is designed to perform I/O in parallel and is more performant than pandas for operations with multiple files or large files.
%%time
import dask
import dask.dataframe as dd
ddf = dd.read_csv(co_sites, parse_dates=['DATE'])
ddf
CPU times: user 462 ms, sys: 123 ms, total: 585 ms
Wall time: 849 ms
| DATE | ID | YEAR | MONTH | DAY | TMAX | TMAX_FLAGS | TMIN | TMIN_FLAGS | PRCP | PRCP_FLAGS | SNOW | SNOW_FLAGS | SNWD | SNWD_FLAGS | WT16 | WT16_FLAGS | WT14 | WT14_FLAGS | WT18 | WT18_FLAGS | TOBS | TOBS_FLAGS | WT01 | WT01_FLAGS | WT08 | WT08_FLAGS | WT03 | WT03_FLAGS | WT04 | WT04_FLAGS | WT06 | WT06_FLAGS | WT05 | WT05_FLAGS | DAPR | DAPR_FLAGS | MDPR | MDPR_FLAGS | WT11 | WT11_FLAGS | SN02 | SN02_FLAGS | SX02 | SX02_FLAGS | SN32 | SN32_FLAGS | SX32 | SX32_FLAGS | SN52 | SN52_FLAGS | SX52 | SX52_FLAGS | LATITUDE | LONGITUDE | ELEVATION | STATE | STATION | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| npartitions=66 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| datetime64[ns] | object | int64 | int64 | int64 | float64 | object | float64 | object | float64 | object | float64 | object | float64 | object | float64 | object | float64 | object | float64 | object | float64 | object | float64 | object | float64 | object | float64 | object | float64 | object | float64 | object | float64 | object | float64 | object | float64 | object | float64 | object | float64 | object | float64 | object | float64 | object | float64 | object | float64 | object | float64 | object | float64 | float64 | float64 | object | object | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
ddf.TMAX.mean()
dd.Scalar<series-..., dtype=float64>
Notice that the representation of the DataFrame object contains no data just headers and datatypes. Why?
Lazy Evaluation#
Similar to Dask Arrays, Dask DataFrames are lazy. Here the data has not yet been read into the dataframe yet (a.k.a. lazy evaluation).
Dask just construct the task graph of the computation but it will “evaluate” them only when necessary.
So how does Dask know the name and dtype of each column?
Dask has just read the start of the first file and infers the column names and dtypes.
Unlike pandas.read_csv that reads in all files before inferring data types, dask.dataframe.read_csv only reads in a sample from the beginning of the file (or first file if using a glob). The column names and dtypes are then enforced when reading the specific partitions (Dask can make mistakes on these inferences if there is missing or misleading data in the early rows).
Let’s take a look at the start of our dataframe:
ddf.head()
| DATE | ID | YEAR | MONTH | DAY | TMAX | TMAX_FLAGS | TMIN | TMIN_FLAGS | PRCP | ... | SX32_FLAGS | SN52 | SN52_FLAGS | SX52 | SX52_FLAGS | LATITUDE | LONGITUDE | ELEVATION | STATE | STATION | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1884-04-01 | USC00035186 | 1884 | 4 | 1 | 289.0 | XX6 | 222.0 | XX6 | 0.0 | ... | XXX | NaN | XXX | NaN | XXX | 35.6042 | -91.2744 | 69.5 | AR | NEWPORT |
| 1 | 1884-04-02 | USC00035186 | 1884 | 4 | 2 | 272.0 | XX6 | 89.0 | XX6 | 0.0 | ... | XXX | NaN | XXX | NaN | XXX | 35.6042 | -91.2744 | 69.5 | AR | NEWPORT |
| 2 | 1884-04-03 | USC00035186 | 1884 | 4 | 3 | 200.0 | XX6 | 22.0 | XX6 | 0.0 | ... | XXX | NaN | XXX | NaN | XXX | 35.6042 | -91.2744 | 69.5 | AR | NEWPORT |
| 3 | 1884-04-04 | USC00035186 | 1884 | 4 | 4 | 244.0 | XX6 | 61.0 | XX6 | 0.0 | ... | XXX | NaN | XXX | NaN | XXX | 35.6042 | -91.2744 | 69.5 | AR | NEWPORT |
| 4 | 1884-04-08 | USC00035186 | 1884 | 4 | 8 | 178.0 | XX6 | 39.0 | XX6 | 0.0 | ... | XXX | NaN | XXX | NaN | XXX | 35.6042 | -91.2744 | 69.5 | AR | NEWPORT |
5 rows × 58 columns
NOTE: Whenever we operate on our dataframe we read through all of our CSV data so that we don’t fill up RAM. Dask will delete intermediate results (like the full pandas DataFrame for each file) as soon as possible. This enables you to handle larger than memory datasets but, repeated computations will have to load all of the data in each time.
Similar data manipulations as pandas.dataframe can be done for dask.dataframes.
For example, let’s find the highest number of snow days in Colorado:
%%time
print ('This site has the highest number of snow days in a year : ')
snowy_days = ddf[ddf['SNOW']>0].groupby(['ID','YEAR']).size()
snowy_days.compute().agg(['idxmax','max'])
This site has the highest number of snow days in a year :
CPU times: user 236 ms, sys: 54.1 ms, total: 290 ms
Wall time: 4.62 s
idxmax (USC00052281, 1983)
max 102
dtype: object
Nice, but what did Dask do?#
# Requires ipywidgets
snowy_days.dask
HighLevelGraph
HighLevelGraph with 6 layers and 331 keys from all layers.
Layer1: read-csv
read-csv-5f2d7502b7b2ca095255b707b5fa92b8
|
Layer2: getitem
getitem-51dd26ae1d79cd0e667caf194aa506cd
|
Layer3: gt
gt-4dc2c28c5c6a8a61c8e747e16a43b818
|
Layer4: getitem
getitem-92164db053c94adfa8b699f100d8db26
|
Layer5: dataframe-groupby-size-chunk
dataframe-groupby-size-chunk-b11331e4b541606fb356e0ef3871f13d-785f1ed81a41cfe4eeefc53d30e1ed67
|
Layer6: dataframe-groupby-size-agg
dataframe-groupby-size-agg-b11331e4b541606fb356e0ef3871f13d
|
You can also view the underlying task graph using .visualize():
#graph is too large
snowy_days.visualize()
2023-02-05 14:09:25,213 - distributed.client - ERROR - Failed to reconnect to scheduler after 30.00 seconds, closing client
Use .compute wisely!#
.persist or caching#
Sometimes you might want your computers to keep intermediate results in memory, if it fits in the memory.
The .persist() method can be used to “cache” data and tell Dask what results to keep around. You should only use .persist() with any data or computation that fits in memory.
For example, if we want to only do analysis on a subset of data (for example snow days at Boulder site):
boulder_snow = ddf[(ddf['SNOW']>0)&(ddf['ID']=='USC00050848')]
%%time
tmax = boulder_snow.TMAX.mean().compute()
tmin = boulder_snow.TMIN.mean().compute()
print (tmin, tmax)
-74.82074711099168 37.419103836866114
CPU times: user 23.8 s, sys: 1.94 s, total: 25.7 s
Wall time: 14.6 s
boulder_snow = ddf[(ddf['SNOW']>0)&(ddf['ID']=='USC00050848')].persist()
%%time
tmax = boulder_snow.TMAX.mean().compute()
tmin = boulder_snow.TMIN.mean().compute()
print (tmin, tmax)
-74.82074711099168 37.419103836866114
CPU times: user 51.9 ms, sys: 4.82 ms, total: 56.7 ms
Wall time: 53.7 ms
As you can see the analysis on this persisted data is much faster because we are not repeating the loading and selecting.
Dask DataFrames Best Practices#
Use pandas (when you can)#
For data that fits into RAM, pandas can often be easier and more efficient to use than Dask DataFrame. However, Dask DataFrame is a powerful tool for larger-than-memory datasets.
When the data is still larger than memory, Dask DataFrame can be used to reduce the larger datasets to a manageable level that pandas can handle. Next, use pandas at that point.
Avoid Full-Data Shuffling#
Some operations are more expensive to compute in a parallel setting than if they are in-memory on a single machine (for example, set_index or merge). In particular, shuffling operations that rearrange data can become very communication intensive.
pandas performance tips#
pandas performance tips such as using vectorized operations also apply to Dask DataFrames. See Modern Pandas notebook for more tips on better performance with pandas.
Check Partition Size#
Similar to chunks, partitions should be small enough that they fit in the memory, but large enough to avoid that the communication overhead.
blocksize#
The number of partitions can be set using the
blocksizeargument. If none is given, the number of partitions/blocksize is calculated depending on the available memory and the number of cores on a machine up to a max of 64 MB. As we increase the blocksize, the number of partitions (calculated by Dask) will decrease. This is especially important when reading one large csv file.
As a good rule of thumb, you should aim for partitions that have around 100MB of data each.
Smart use of .compute()#
Try avoiding running .compute() operation as long as possible. Dask works best when users avoid computation until results are needed. The .compute() command informs Dask to trigger computations on the Dask DataFrame.
As shown in the above example, the intermediate results can also be shared by calling .compute() only once.
Close your local Dask Cluster#
It is always a good practice to close the Dask cluster you created.
client.shutdown()
Summary#
In this notebook, we have learned about:
Dask DataFrame concept and component.
When to use and when to avoid Dask DataFrames?
How to use Dask DataFrame?
Some best practices around Dask DataFrames.
Resources and references#
Reference
Ask for help
dasktag on Stack Overflow, for usage questionsgithub discussions: dask for general, non-bug, discussion, and usage questions
github issues: dask for bug reports and feature requests


