Section 1: Introduction¶
Python package imports and useful function definitions
# Import
import intake
import numpy as np
import xarray as xr
import nc_time_axis
import osimport dask
from dask_jobqueue import PBSCluster
from dask.distributed import Client
from dask.distributed import performance_report# Set up your sratch folder path
username = os.environ["USER"]
glade_scratch = "/glade/derecho/scratch/" + username
print(glade_scratch)/glade/derecho/scratch/harshah
# catalog_url = 'https://data-osdf.gdex.ucar.edu/ncar-gdex/d010092/catalogs/d010092-osdf.json'
catalog_url = 'https://stratus.gdex.ucar.edu/d010092/catalogs/d010092-https.json' #NCAR's Object store# GMST function ###
# calculate global means
def get_lat_name(ds):
for lat_name in ['lat', 'latitude']:
if lat_name in ds.coords:
return lat_name
raise RuntimeError("Couldn't find a latitude coordinate")
def global_mean(ds):
lat = ds[get_lat_name(ds)]
weight = np.cos(np.deg2rad(lat))
weight /= weight.mean()
other_dims = set(ds.dims) - {'time','member_id'}
return (ds * weight).mean(other_dims)Section 2: Set up a Dask Cluster on Casper¶
Setting up a dask cluster. You will need an NCAR HPC account to do this.
# Create a PBS cluster object
cluster = PBSCluster(
job_name = 'dask-wk25',
cores = 1,
memory = '8GiB',
processes = 1,
local_directory = glade_scratch+'/dask/spill/',
log_directory = glade_scratch + '/dask/logs/',
resource_spec = 'select=1:ncpus=1:mem=8GB',
queue = 'casper',
walltime = '5:00:00',
interface = 'ext'
)# Create the client to load the Dashboard
client = Client(cluster)n_workers = 5
cluster.scale(n_workers)
client.wait_for_workers(n_workers = n_workers)
clusterLoading...
Section 3: Data Loading¶
Load CESM2 LENS zarr data from GDEX using an intake-ESM catalog
For more details regarding the dataset. See, https://
gdex .ucar .edu /datasets /d010092 /#
# Open collection description file using intake
col = intake.open_esm_datastore(catalog_url)
colLoading...
cesm_temp = col.search(variable ='TREFHT', frequency ='monthly')
cesm_tempLoading...
cesm_temp.df['path'].valuesarray(['https://stratus.gdex.ucar.edu/d010092/atm/monthly/cesm2LE-historical-cmip6-TREFHT.zarr',
'https://stratus.gdex.ucar.edu/d010092/atm/monthly/cesm2LE-historical-smbb-TREFHT.zarr',
'https://stratus.gdex.ucar.edu/d010092/atm/monthly/cesm2LE-ssp370-cmip6-TREFHT.zarr',
'https://stratus.gdex.ucar.edu/d010092/atm/monthly/cesm2LE-ssp370-smbb-TREFHT.zarr'],
dtype=object)dsets_cesm = cesm_temp.to_dataset_dict()
--> The keys in the returned dictionary of datasets are constructed as follows:
'component.experiment.frequency.forcing_variant'
Loading...
Loading...
dsets_cesm.keys()dict_keys(['atm.historical.monthly.cmip6', 'atm.ssp370.monthly.cmip6', 'atm.historical.monthly.smbb', 'atm.ssp370.monthly.smbb'])historical_cmip6 = dsets_cesm['atm.historical.monthly.cmip6']
future_cmip6 = dsets_cesm['atm.ssp370.monthly.cmip6']future_cmip6 Loading...
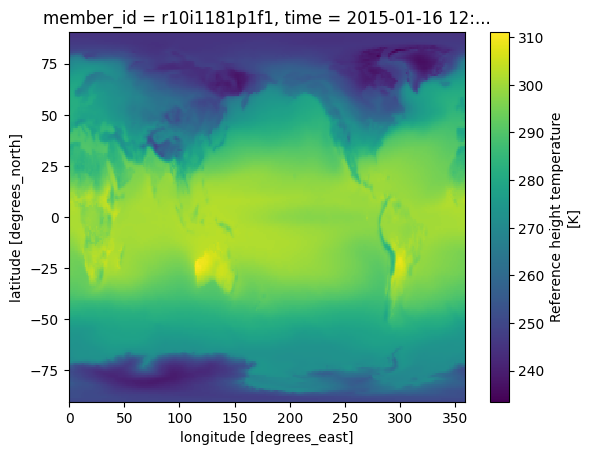
Make a quick plot to check data transfer¶
%%time
future_cmip6.TREFHT.isel(member_id=0,time=0).plot()CPU times: user 161 ms, sys: 8.8 ms, total: 170 ms
Wall time: 1.11 s
Section 4: Data Analysis¶
Perform the Global Mean Surface Temperature computation
Merge datasets and compute Global Mean surrface temperature anomaly¶
Warning! This section takes about a min to run!
Config: 3 dask workers with 8GiB memory.
merge_ds_cmip6 = xr.concat([historical_cmip6, future_cmip6], dim='time')
# merge_ds_cmip6 = merge_ds_cmip6.dropna(dim='member_id')
merge_ds_cmip6 = merge_ds_cmip6.TREFHT
merge_ds_cmip6Loading...
Compute (spatially weighted) Global Mean¶
ds_cmip6_annual = merge_ds_cmip6.resample(time='YS').mean()
ds_cmip6_annualLoading...
%%time
gmst_cmip6 = global_mean(ds_cmip6_annual)
gmst_cmip6 = gmst_cmip6.rename('gmst')
gmst_cmip6CPU times: user 460 ms, sys: 64.1 ms, total: 524 ms
Wall time: 572 ms
Loading...
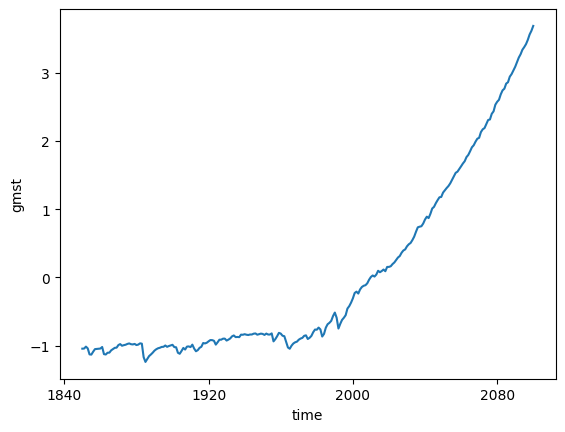
Compute anomaly and plot¶
gmst_cmip6_ano = gmst_cmip6 - gmst_cmip6.mean()
gmst_cmip6_anoLoading...
gmst_cmip6_ano = gmst_cmip6_ano.compute()%%time
gmst_cmip6_ano.mean(dim='member_id').plot()CPU times: user 25.8 ms, sys: 407 μs, total: 26.2 ms
Wall time: 30.4 ms
cluster.close()