specx_anal#
Warning
This is not meant to be a standalone notebook. This notebook is part of the process we have for adding entries to the NCL Index and is not meant to be used as tutorial or example code.
import xarray as xr
import geocat.datafiles as gcd
import pandas as pd
import scipy
import matplotlib.pyplot as plt
import numpy as np
NCL code#
;*************************************************
; spec_1.ncl
;
; Concepts illustrated:
; - Calculating and plotting spectra
;************************************************
;
; These files are loaded by default in NCL V6.2.0 and newer
; load "$NCARG_ROOT/lib/ncarg/nclscripts/csm/gsn_code.ncl"
; load "$NCARG_ROOT/lib/ncarg/nclscripts/csm/gsn_csm.ncl"
;************************************************
begin
;************************************************
; variable and file handling
;************************************************
fn = "SOI_Darwin.nc" ; define filename
in = addfile(fn,"r") ; open netcdf file
soi = in->DSOI ; get data
;************************************************
; set function arguments
;************************************************
; detrending opt: 0=>remove mean 1=>remove mean and detrend
d = 0
; smoothing periodogram: (0 <= sm <= ??.) should be at least 3 and odd
sm = 7
; percent tapered: (0.0 <= pct <= 1.0) 0.10 common.
pct = 0.10
;************************************************
; calculate spectrum
;************************************************
spec = specx_anal(soi,d,sm,pct)
;************************************************
; plotting
;************************************************
wks = gsn_open_wks("png","spec") ; send graphics to PNG file
res = True ; plot mods desired
res@tiMainString = "SOI" ; title
res@tiXAxisString = "Frequency (cycles/month)" ; xaxis
res@tiYAxisString = "Variance" ; yaxis
plot=gsn_csm_xy(wks,spec@frq,spec@spcx,res) ; create plot
;***********************************************
end
Python Functionality#
soi_darwin = xr.open_dataset(gcd.get('netcdf_files/SOI_Darwin.nc'))
soi_darwin
<xarray.Dataset> Size: 22kB
Dimensions: (time: 1404)
Coordinates:
* time (time) int32 6kB 0 1 2 3 4 5 6 ... 1398 1399 1400 1401 1402 1403
Data variables:
date (time) float64 11kB ...
DSOI (time) float32 6kB ...
Attributes:
title: Darwin Southern Oscillation Index
source: Climate Analysis Section, NCAR
history: \nDSOI = - Normalized Darwin\nNormalized sea level pressu...
creation_date: Tue Mar 30 09:29:20 MST 1999
references: Trenberth, Mon. Wea. Rev: 2/1984
time_span: 1882 - 1998
conventions: Nonestart_date = pd.Timestamp('1882-01-01')
dates = [
start_date + pd.DateOffset(months=int(month))
for month in soi_darwin.indexes['time']
]
datetime_index = pd.DatetimeIndex(dates)
datetime_index
DatetimeIndex(['1882-01-01', '1882-02-01', '1882-03-01', '1882-04-01',
'1882-05-01', '1882-06-01', '1882-07-01', '1882-08-01',
'1882-09-01', '1882-10-01',
...
'1998-03-01', '1998-04-01', '1998-05-01', '1998-06-01',
'1998-07-01', '1998-08-01', '1998-09-01', '1998-10-01',
'1998-11-01', '1998-12-01'],
dtype='datetime64[us]', length=1404, freq=None)
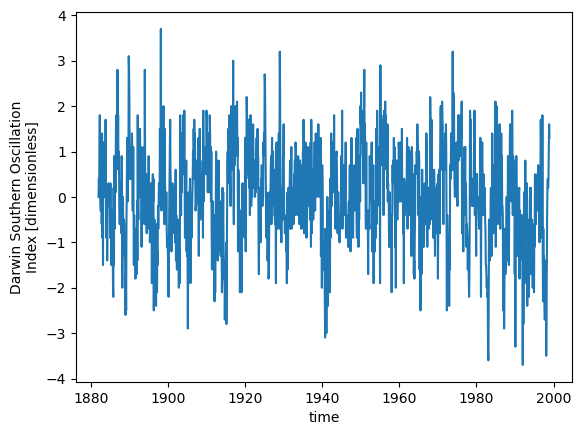
soi = soi_darwin.DSOI
soi['time'] = datetime_index
soi.plot();
soi_detrended = scipy.signal.detrend(soi, type='constant')
percent_taper = 0.1
tukey_window = scipy.signal.windows.tukey(
len(soi_detrended), alpha=percent_taper, sym=False
) # generates a periodic window
soi_tapered = soi_detrended * tukey_window
freq_soi, psd_soi = scipy.signal.periodogram(
soi_tapered,
fs=1, # sample monthly
detrend=False,
)
k = 7 # define smoothing constant
kernel = np.ones(7) # Create a Daniel smoothing kernel
kernel[0] = (
0.5 # "Modify" kernel by making the endpoints have half the weight of the interior points
)
kernel[-1] = 0.5
kernel = kernel / kernel.sum()
smoothed_psd = scipy.signal.convolve(
psd_soi, kernel, mode='same'
) # Sets output array as the same length as the first input
variance = np.var(soi_tapered, ddof=1)
df = freq_soi[1] - freq_soi[0] # Frequency step
# Create an array to adjust contributions of endpoints
frac = np.ones_like(freq_soi)
frac[0] = 0.5
frac[-1] = 0.5
current_area = np.sum(
smoothed_psd * df * frac
) # Calculate the current area under the curve
normalization_factor = variance / current_area # Find the factor to adjust this area
normalized_psd = (
smoothed_psd * normalization_factor
) # Apply the normalization factor to the smoothed power spectrum
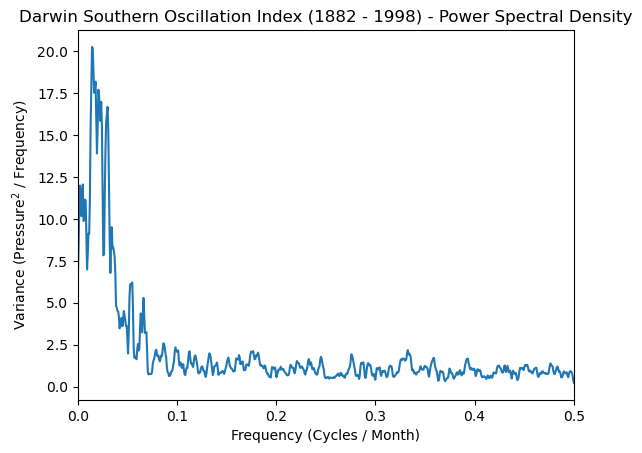
plt.plot(freq_soi, normalized_psd)
plt.xlabel('Frequency (Cycles / Month)')
plt.ylabel('Variance (Pressure$^2$ / Frequency)')
plt.title('Darwin Southern Oscillation Index (1882 - 1998) - Power Spectral Density')
plt.xlim([0, 0.5]);
Comparison#
ncl = xr.open_dataset(gcd.get('applications_files/ncl_outputs/spec_1_output.nc')).spec
frq = ncl.frq
spcx = ncl.spcx
Downloading file 'applications_files/ncl_outputs/spec_1_output.nc' from 'https://github.com/NCAR/geocat-datafiles/raw/main/applications_files/ncl_outputs/spec_1_output.nc' to '/home/runner/.cache/geocat'.
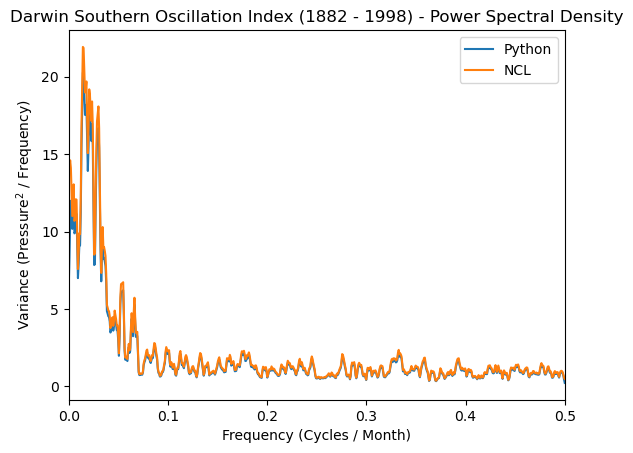
plt.plot(freq_soi, normalized_psd, label='Python')
plt.plot(frq, spcx, label='NCL')
plt.xlabel('Frequency (Cycles / Month)')
plt.ylabel('Variance (Pressure$^2$ / Frequency)')
plt.title('Darwin Southern Oscillation Index (1882 - 1998) - Power Spectral Density')
plt.xlim([0, 0.5])
plt.legend();
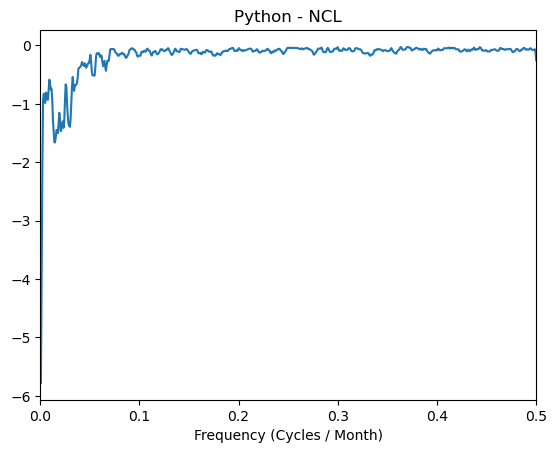
plt.plot(frq, normalized_psd[1:] - spcx)
plt.xlim([0, 0.5])
plt.title('Python - NCL')
plt.xlabel('Frequency (Cycles / Month)');
Comparison without Tapering#
It is important to remember that periodograms are estimates of a spectra, and the recommended math for performing this analysis, as well as for the various window operations, has changed over the decades since NCL was written.
We have demonstrated to the best of our ability how to follow the decisions made by NCL developers and have results similar enough to be confident that this approach in Python is correct.
However, in the interest of tracking down the differences we will investigate the output from Python and NCL without tapering.
Because Tukey tapering can refer to split-bell-cosine or cosine tapering, this is the step of our workflow where NCL’s decisions are the least clear.
ncl_notaper = xr.open_dataset(
gcd.get('applications_files/ncl_outputs/spec_1_output_notaper.nc')
).spec
spcx_nt = ncl_notaper.spcx
Downloading file 'applications_files/ncl_outputs/spec_1_output_notaper.nc' from 'https://github.com/NCAR/geocat-datafiles/raw/main/applications_files/ncl_outputs/spec_1_output_notaper.nc' to '/home/runner/.cache/geocat'.
# Compute Periodogram without tapering and with mean removed
freq_soi, psd_soi_nt = scipy.signal.periodogram(
soi_detrended,
fs=1, # sample monthly
detrend=False,
)
# Smooth with the same modified Daniel kernel as before
smoothed_psd_nt = scipy.signal.convolve(psd_soi_nt, kernel, mode='same')
# Normalize
variance = np.var(soi_detrended, ddof=1)
current_area = np.sum(
smoothed_psd_nt * df * frac
) # Calculate the current area under the curve
normalization_factor = variance / current_area # Find the factor to adjust this area
normalized_psd_nt = (
smoothed_psd_nt * normalization_factor
) # Apply the normalization factor to the smoothed power spectrum
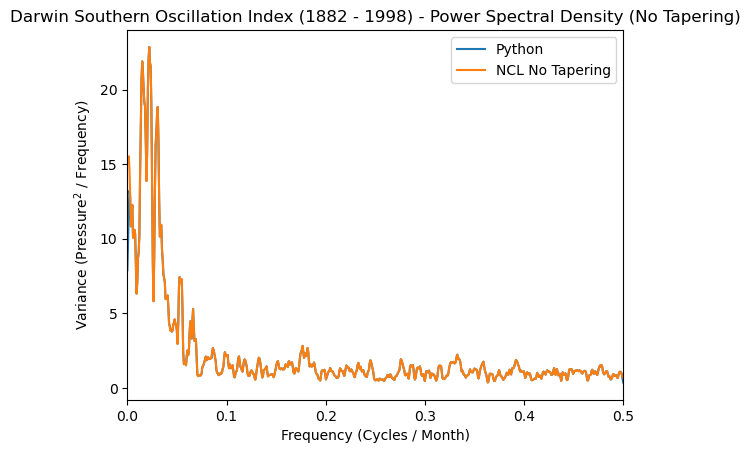
plt.plot(freq_soi, normalized_psd_nt, label='Python')
plt.plot(frq, spcx_nt, label='NCL No Tapering')
plt.xlabel('Frequency (Cycles / Month)')
plt.ylabel('Variance (Pressure$^2$ / Frequency)')
plt.title(
'Darwin Southern Oscillation Index (1882 - 1998) - Power Spectral Density (No Tapering)'
)
plt.xlim([0, 0.5])
plt.legend();
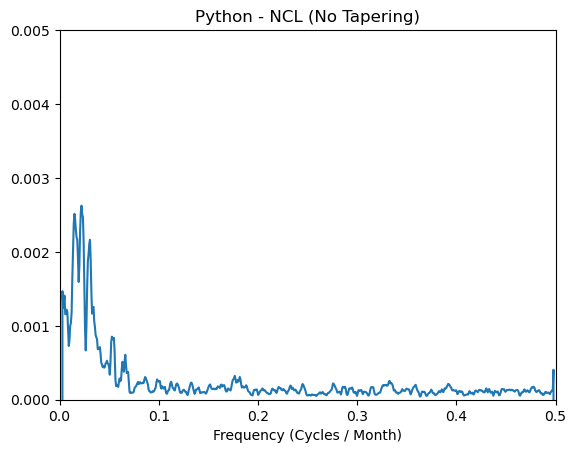
plt.plot(frq, normalized_psd_nt[1:] - spcx_nt)
plt.xlim([0, 0.5])
plt.title('Python - NCL (No Tapering)')
plt.ylim([-0.00, 0.005])
plt.xlabel('Frequency (Cycles / Month)');