# Display output of plots directly in Notebook
%matplotlib inline
import warnings
warnings.filterwarnings("ignore")
import intake
import numpy as np
import pandas as pd
import xarray as xr
import s3fs
import seaborn as snsimport dask
from dask_jobqueue import PBSCluster
from dask.distributed import Client
from dask.distributed import performance_reportrda_scratch = '/gpfs/csfs1/collections/rda/scratch/harshah'# Create a PBS cluster object
cluster = PBSCluster(
job_name = 'dask-wk24-hpc',
cores = 1,
memory = '8GiB',
processes = 1,
local_directory = rda_scratch+'/dask/spill',
resource_spec = 'select=1:ncpus=1:mem=8GB',
queue = 'casper',
walltime = '2:00:00',
#interface = 'ib0'
interface = 'ext'
)# Open collection description file using intake
catalog_url = 'https://ncar-cesm-lens.s3-us-west-2.amazonaws.com/catalogs/aws-cesm1-le.json'
col = intake.open_esm_datastore(catalog_url)
colLoading...
# Use s3fs
s3 = s3fs.S3FileSystem(anon=True)
# List top-level directories and files in the 'ncar-cesm2-lens' bucket
bucket_contents = s3.ls('ncar-cesm2-lens/atm/monthly')
print(bucket_contents)['ncar-cesm2-lens/atm/monthly/', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-FLNS.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-FLNSC.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-FLUT.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-FSNS.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-FSNSC.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-FSNTOA.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-ICEFRAC.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-LHFLX.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-PRECC.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-PRECL.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-PRECSC.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-PRECSL.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-PS.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-PSL.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-Q.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-SHFLX.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-T.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-TMQ.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-TREFHT.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-TREFHTMN.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-TREFHTMX.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-TS.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-U.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-Z3.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-FLNS.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-FLNSC.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-FLUT.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-FSNS.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-FSNSC.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-FSNTOA.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-ICEFRAC.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-LHFLX.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-PRECC.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-PRECL.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-PRECSC.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-PRECSL.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-PS.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-PSL.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-Q.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-SHFLX.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-T.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-TMQ.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-TREFHT.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-TREFHTMN.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-TREFHTMX.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-TS.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-U.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-historical-smbb-Z3.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-cmip6-FLNS.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-cmip6-FLNSC.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-cmip6-FLUT.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-cmip6-FSNS.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-cmip6-FSNSC.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-cmip6-FSNTOA.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-cmip6-ICEFRAC.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-cmip6-LHFLX.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-cmip6-PRECC.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-cmip6-PRECL.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-cmip6-PRECSC.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-cmip6-PRECSL.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-cmip6-PS.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-cmip6-PSL.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-cmip6-Q.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-cmip6-SHFLX.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-cmip6-T.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-cmip6-TMQ.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-cmip6-TREFHT.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-cmip6-TREFHTMN.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-cmip6-TREFHTMX.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-cmip6-TS.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-cmip6-U.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-cmip6-Z3.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-smbb-FLNS.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-smbb-FLNSC.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-smbb-FLUT.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-smbb-FSNS.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-smbb-FSNSC.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-smbb-FSNTOA.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-smbb-ICEFRAC.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-smbb-LHFLX.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-smbb-PRECC.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-smbb-PRECL.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-smbb-PRECSC.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-smbb-PRECSL.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-smbb-PS.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-smbb-PSL.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-smbb-Q.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-smbb-SHFLX.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-smbb-T.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-smbb-TMQ.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-smbb-TREFHT.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-smbb-TREFHTMN.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-smbb-TREFHTMX.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-smbb-TS.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-smbb-U.zarr', 'ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-smbb-Z3.zarr']
# Get data for the members which use the cmip6 prescription for biomass burning
store_hist_cmip6 = s3fs.S3Map(root='ncar-cesm2-lens/atm/monthly/cesm2LE-historical-cmip6-TREFHT.zarr', s3=s3)
hist_cmip6 = xr.open_zarr(store_hist_cmip6)['TREFHT']
hist_cmip6Loading...
# Get the corresponding future data which uses ssp370 pathway
store_ssp370_cmip6 = s3fs.S3Map(root='ncar-cesm2-lens/atm/monthly/cesm2LE-ssp370-cmip6-TREFHT.zarr', s3=s3)
ssp370_cmip6 = xr.open_zarr(store_ssp370_cmip6)['TREFHT']
ssp370_cmip6Loading...
Merge the data¶
ds_cmip6 = xr.concat([hist_cmip6,ssp370_cmip6],dim='time')
ds_cmip6Loading...
# GMST function ###
# calculate global means
def get_lat_name(ds):
for lat_name in ['lat', 'latitude']:
if lat_name in ds.coords:
return lat_name
raise RuntimeError("Couldn't find a latitude coordinate")
def global_mean(ds):
lat = ds[get_lat_name(ds)]
weight = np.cos(np.deg2rad(lat))
weight /= weight.mean()
other_dims = set(ds.dims) - {'time','member_id'}
return (ds * weight).mean(other_dims)client = Client(cluster)
clientLoading...
cluster.scale(8)
clusterLoading...
Calculate GMST¶
Now compute (spatially weighted) Global Mean¶
ds_cmip6_annual = ds_cmip6.resample(time='AS').mean()
ds_cmip6_annualLoading...
%%time
gmst_cmip6 = global_mean(ds_cmip6_annual)
gmst_cmip6 = gmst_cmip6.rename('gmst')
gmst_cmip6Loading...
# %%time
# gmst_cmip6.to_dataset().to_zarr(rda_scratch + 'cesm2/gmst_cmip6.zarr',mode='w')%%time
gmst_cmip6 = xr.open_zarr(rda_scratch + 'cesm2/gmst_cmip6.zarr').gmst
gmst_cmip6Loading...
Compute anomaly and turn the result into a data frame¶
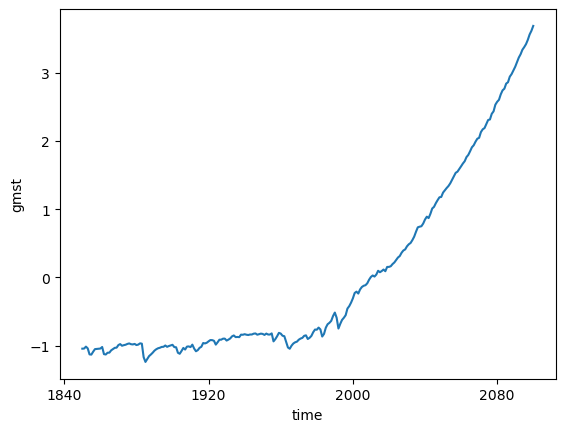
gmsta_cmip6 = gmst_cmip6 - gmst_cmip6.mean()%%time
gmsta_cmip6.mean(dim='member_id').plot()CPU times: user 24.6 s, sys: 1.48 s, total: 26.1 s
Wall time: 4min 8s
gmsta_cmip6_df = gmsta_cmip6.to_dataframe().reset_index()gmsta_cmip6_dfLoading...
%%time
sns.relplot(data=gmsta_cmip6_df,
x="time", y="gmst", hue='member_id',
kind="line", ci="sd", aspect=2)---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
File /glade/work/harshah/conda-envs/zarr_experiments/lib/python3.11/site-packages/matplotlib/axis.py:1769, in Axis.convert_units(self, x)
1768 try:
-> 1769 ret = self.converter.convert(x, self.units, self)
1770 except Exception as e:
File /glade/work/harshah/conda-envs/zarr_experiments/lib/python3.11/site-packages/nc_time_axis/__init__.py:556, in NetCDFTimeConverter.convert(cls, value, unit, axis)
555 if not isinstance(first_value, (CalendarDateTime, cftime.datetime)):
--> 556 raise ValueError(
557 "The values must be numbers or instances of "
558 '"nc_time_axis.CalendarDateTime" or '
559 '"cftime.datetime".'
560 )
562 if isinstance(first_value, CalendarDateTime):
ValueError: The values must be numbers or instances of "nc_time_axis.CalendarDateTime" or "cftime.datetime".
The above exception was the direct cause of the following exception:
ConversionError Traceback (most recent call last)
File <timed eval>:1
File /glade/work/harshah/conda-envs/zarr_experiments/lib/python3.11/site-packages/seaborn/relational.py:838, in relplot(data, x, y, hue, size, style, units, weights, row, col, col_wrap, row_order, col_order, palette, hue_order, hue_norm, sizes, size_order, size_norm, markers, dashes, style_order, legend, kind, height, aspect, facet_kws, **kwargs)
829 g = FacetGrid(
830 data=full_data.dropna(axis=1, how="all"),
831 **grid_kws,
(...)
834 **facet_kws
835 )
837 # Draw the plot
--> 838 g.map_dataframe(func, **plot_kws)
840 # Label the axes, using the original variables
841 # Pass "" when the variable name is None to overwrite internal variables
842 g.set_axis_labels(variables.get("x") or "", variables.get("y") or "")
File /glade/work/harshah/conda-envs/zarr_experiments/lib/python3.11/site-packages/seaborn/axisgrid.py:825, in FacetGrid.map_dataframe(self, func, *args, **kwargs)
822 kwargs["data"] = data_ijk
824 # Draw the plot
--> 825 self._facet_plot(func, ax, args, kwargs)
827 # For axis labels, prefer to use positional args for backcompat
828 # but also extract the x/y kwargs and use if no corresponding arg
829 axis_labels = [kwargs.get("x", None), kwargs.get("y", None)]
File /glade/work/harshah/conda-envs/zarr_experiments/lib/python3.11/site-packages/seaborn/axisgrid.py:854, in FacetGrid._facet_plot(self, func, ax, plot_args, plot_kwargs)
852 plot_args = []
853 plot_kwargs["ax"] = ax
--> 854 func(*plot_args, **plot_kwargs)
856 # Sort out the supporting information
857 self._update_legend_data(ax)
File /glade/work/harshah/conda-envs/zarr_experiments/lib/python3.11/site-packages/seaborn/relational.py:515, in lineplot(data, x, y, hue, size, style, units, weights, palette, hue_order, hue_norm, sizes, size_order, size_norm, dashes, markers, style_order, estimator, errorbar, n_boot, seed, orient, sort, err_style, err_kws, legend, ci, ax, **kwargs)
512 color = kwargs.pop("color", kwargs.pop("c", None))
513 kwargs["color"] = _default_color(ax.plot, hue, color, kwargs)
--> 515 p.plot(ax, kwargs)
516 return ax
File /glade/work/harshah/conda-envs/zarr_experiments/lib/python3.11/site-packages/seaborn/relational.py:276, in _LinePlotter.plot(self, ax, kws)
268 # TODO How to handle NA? We don't want NA to propagate through to the
269 # estimate/CI when some values are present, but we would also like
270 # matplotlib to show "gaps" in the line when all values are missing.
(...)
273
274 # Loop over the semantic subsets and add to the plot
275 grouping_vars = "hue", "size", "style"
--> 276 for sub_vars, sub_data in self.iter_data(grouping_vars, from_comp_data=True):
278 if self.sort:
279 sort_vars = ["units", orient, other]
File /glade/work/harshah/conda-envs/zarr_experiments/lib/python3.11/site-packages/seaborn/_base.py:902, in VectorPlotter.iter_data(self, grouping_vars, reverse, from_comp_data, by_facet, allow_empty, dropna)
899 grouping_vars = [var for var in grouping_vars if var in self.variables]
901 if from_comp_data:
--> 902 data = self.comp_data
903 else:
904 data = self.plot_data
File /glade/work/harshah/conda-envs/zarr_experiments/lib/python3.11/site-packages/seaborn/_base.py:1000, in VectorPlotter.comp_data(self)
995 if var in self.var_levels:
996 # TODO this should happen in some centralized location
997 # it is similar to GH2419, but more complicated because
998 # supporting `order` in categorical plots is tricky
999 orig = orig[orig.isin(self.var_levels[var])]
-> 1000 comp = pd.to_numeric(converter.convert_units(orig)).astype(float)
1001 transform = converter.get_transform().transform
1002 parts.append(pd.Series(transform(comp), orig.index, name=orig.name))
File /glade/work/harshah/conda-envs/zarr_experiments/lib/python3.11/site-packages/matplotlib/axis.py:1771, in Axis.convert_units(self, x)
1769 ret = self.converter.convert(x, self.units, self)
1770 except Exception as e:
-> 1771 raise munits.ConversionError('Failed to convert value(s) to axis '
1772 f'units: {x!r}') from e
1773 return ret
ConversionError: Failed to convert value(s) to axis units: 0 1850-01-01 00:00:00
1 1851-01-01 00:00:00
2 1852-01-01 00:00:00
3 1853-01-01 00:00:00
4 1854-01-01 00:00:00
...
12545 2096-01-01 00:00:00
12546 2097-01-01 00:00:00
12547 2098-01-01 00:00:00
12548 2099-01-01 00:00:00
12549 2100-01-01 00:00:00
Name: x, Length: 12550, dtype: object