import glob
import re
import matplotlib as plt
import numpy as np
import scipy as sp
import xarray as xr
import intake
import intake_esm
import pandas as pd######## File paths ################
lustre_scratch = "/lustre/desc1/scratch/harshah"
gdex_data = "/gdex/data/special_projects/pythia_2024"
annual_means = gdex_data + '/annual_means/'
zarr_path = gdex_data + "/tas_zarr/"
#########
gdex_url = 'https://data.gdex.ucar.edu/'
era5_catalog = gdex_url + 'special_projects/pythia_2024/pythia_intake_catalogs/era5_catalog.json'
#
##########
print(era5_catalog)https://data.gdex.ucar.edu/special_projects/pythia_2024/pythia_intake_catalogs/era5_catalog.json
USE_PBS_SCHEDULER = True# Create a PBS cluster object
def get_pbs_cluster():
""" Create cluster through dask_jobqueue.
"""
from dask_jobqueue import PBSCluster
cluster = PBSCluster(
job_name = 'dask-osdf-24',
cores = 1,
memory = '4GiB',
processes = 1,
local_directory = lustre_scratch + '/dask/spill',
log_directory = lustre_scratch + '/dask/logs/',
resource_spec = 'select=1:ncpus=1:mem=4GB',
queue = 'casper',
walltime = '3:00:00',
#interface = 'ib0'
interface = 'ext'
)
return cluster
def get_gateway_cluster():
""" Create cluster through dask_gateway
"""
from dask_gateway import Gateway
gateway = Gateway()
cluster = gateway.new_cluster()
cluster.adapt(minimum=2, maximum=4)
return cluster
def get_local_cluster():
""" Create cluster using the Jupyter server's resources
"""
from distributed import LocalCluster, performance_report
cluster = LocalCluster()
cluster.scale(6)
return cluster# Obtain dask cluster in one of three ways
if USE_PBS_SCHEDULER:
cluster = get_pbs_cluster()
elif USE_DASK_GATEWAY:
cluster = get_gateway_cluster()
else:
cluster = get_local_cluster()
# Connect to cluster
from distributed import Client
client = Client(cluster)/glade/u/home/harshah/venvs/osdf/lib/python3.10/site-packages/distributed/node.py:187: UserWarning: Port 8787 is already in use.
Perhaps you already have a cluster running?
Hosting the HTTP server on port 41997 instead
warnings.warn(
# Scale the cluster and display cluster dashboard URL
n_workers =5
cluster.scale(n_workers)
client.wait_for_workers(n_workers = n_workers)
clusterLoading...
%pip show intake-esmName: intake-esm
Version: 2025.2.3
Summary: An intake plugin for parsing an Earth System Model (ESM) catalog and loading netCDF files and/or Zarr stores into Xarray datasets.
Home-page: https://intake-esm.readthedocs.io
Author:
Author-email:
License: Apache Software License 2.0
Location: /glade/u/home/harshah/venvs/osdf/lib/python3.10/site-packages
Requires: dask, fastprogress, fsspec, intake, netCDF4, pandas, pydantic, requests, xarray, zarr
Required-by: ecgtools
Note: you may need to restart the kernel to use updated packages.
era5_cat = intake.open_esm_datastore(era5_catalog)
era5_catLoading...
era5_cat.dfLoading...
temp_cat = era5_cat.search(variable='VAR_2T',frequency = 'hourly',year=2000)
temp_catLoading...
# Define the xarray_open_kwargs with a compatible engine, for example, 'scipy'
xarray_open_kwargs = {
'engine': 'h5netcdf',
'chunks': {}, # Specify any chunking if needed
'backend_kwargs': {} # Any additional backend arguments if required
}%%time
dset_temp = temp_cat.to_dataset_dict(xarray_open_kwargs=xarray_open_kwargs)Loading...
dset_temp{'an.sfc': <xarray.Dataset> Size: 36GB
Dimensions: (time: 8784, latitude: 721, longitude: 1440)
Coordinates:
* latitude (latitude) float64 6kB 90.0 89.75 89.5 ... -89.5 -89.75 -90.0
* longitude (longitude) float64 12kB 0.0 0.25 0.5 0.75 ... 359.2 359.5 359.8
* time (time) datetime64[ns] 70kB 2000-01-01 ... 2000-12-31T23:00:00
utc_date (time) int32 35kB dask.array<chunksize=(744,), meta=np.ndarray>
Data variables:
VAR_2T (time, latitude, longitude) float32 36GB dask.array<chunksize=(27, 139, 277), meta=np.ndarray>
Attributes: (12/18)
DATA_SOURCE: ECMWF: https://cds.climate.copernicus.eu...
NETCDF_CONVERSION: CISL RDA: Conversion from ECMWF GRIB1 da...
Conventions: CF-1.6
NETCDF_COMPRESSION: NCO: Precision-preserving compression to...
intake_esm_vars: ['VAR_2T']
intake_esm_attrs:era_id: e5
... ...
intake_esm_attrs:units: K
intake_esm_attrs:year: 2000
intake_esm_attrs:format: nc
intake_esm_attrs:frequency: hourly
intake_esm_attrs:_data_format_: netcdf
intake_esm_dataset_key: an.sfc}temps = dset_temp['an.sfc']temps.VAR_2T.isel(time=0).plot()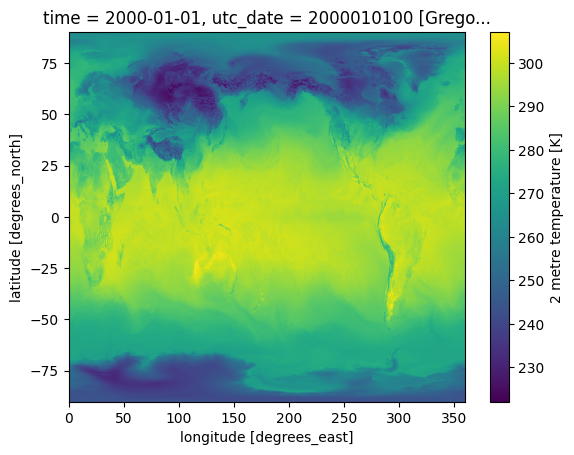
cluster.close()