Writing multiple netCDF files in parallel with xarray and dask#
A typical computation workflow with xarray consists of:
reading one or more netCDF files into an xarray dataset backed by dask using
xr.open_mfdataset()orxr.open_dataset(chunks=...),applying some transformation to the input dataset, and
saving the resulting output to disk in a netCDF file using
xr.to_netcdf().
The last step (3) can easily lead to a large netCDF file (>=10GB in size). As a result, this step can take a very long time to complete (since it is run in serial), and sometimes may hang. So, to avoid these issues one can use one of the lesser-used but helpful xarray capabilities: the xr.save_mfdataset() function. This function allows users to write multiple datasets to disk as netCDF files simultaneously. The xr.save_mfdataset() function signature looks like this:
xr.save_mfdataset(
datasets,
paths,
mode='w',
format=None,
groups=None,
engine=None,
compute=True,
)
Docstring:
Write multiple datasets to disk as netCDF files simultaneously.
Please show me the code#
Package imports#
import numpy as np
import xarray as xr
from distributed import Client, performance_report
xr.__version__
'0.15.1'
client = Client()
client
Client
|
Cluster
|
Load some toy dataset#
ds = xr.tutorial.open_dataset('rasm', chunks={'time': 12})
ds
- time: 36
- x: 275
- y: 205
- time(time)object1980-09-16 12:00:00 ... 1983-08-17 00:00:00
- long_name :
- time
- type_preferred :
- int
array([cftime.DatetimeNoLeap(1980-09-16 12:00:00), cftime.DatetimeNoLeap(1980-10-17 00:00:00), cftime.DatetimeNoLeap(1980-11-16 12:00:00), cftime.DatetimeNoLeap(1980-12-17 00:00:00), cftime.DatetimeNoLeap(1981-01-17 00:00:00), cftime.DatetimeNoLeap(1981-02-15 12:00:00), cftime.DatetimeNoLeap(1981-03-17 00:00:00), cftime.DatetimeNoLeap(1981-04-16 12:00:00), cftime.DatetimeNoLeap(1981-05-17 00:00:00), cftime.DatetimeNoLeap(1981-06-16 12:00:00), cftime.DatetimeNoLeap(1981-07-17 00:00:00), cftime.DatetimeNoLeap(1981-08-17 00:00:00), cftime.DatetimeNoLeap(1981-09-16 12:00:00), cftime.DatetimeNoLeap(1981-10-17 00:00:00), cftime.DatetimeNoLeap(1981-11-16 12:00:00), cftime.DatetimeNoLeap(1981-12-17 00:00:00), cftime.DatetimeNoLeap(1982-01-17 00:00:00), cftime.DatetimeNoLeap(1982-02-15 12:00:00), cftime.DatetimeNoLeap(1982-03-17 00:00:00), cftime.DatetimeNoLeap(1982-04-16 12:00:00), cftime.DatetimeNoLeap(1982-05-17 00:00:00), cftime.DatetimeNoLeap(1982-06-16 12:00:00), cftime.DatetimeNoLeap(1982-07-17 00:00:00), cftime.DatetimeNoLeap(1982-08-17 00:00:00), cftime.DatetimeNoLeap(1982-09-16 12:00:00), cftime.DatetimeNoLeap(1982-10-17 00:00:00), cftime.DatetimeNoLeap(1982-11-16 12:00:00), cftime.DatetimeNoLeap(1982-12-17 00:00:00), cftime.DatetimeNoLeap(1983-01-17 00:00:00), cftime.DatetimeNoLeap(1983-02-15 12:00:00), cftime.DatetimeNoLeap(1983-03-17 00:00:00), cftime.DatetimeNoLeap(1983-04-16 12:00:00), cftime.DatetimeNoLeap(1983-05-17 00:00:00), cftime.DatetimeNoLeap(1983-06-16 12:00:00), cftime.DatetimeNoLeap(1983-07-17 00:00:00), cftime.DatetimeNoLeap(1983-08-17 00:00:00)], dtype=object) - xc(y, x)float64dask.array<chunksize=(205, 275), meta=np.ndarray>
- long_name :
- longitude of grid cell center
- units :
- degrees_east
- bounds :
- xv
Array Chunk Bytes 451.00 kB 451.00 kB Shape (205, 275) (205, 275) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - yc(y, x)float64dask.array<chunksize=(205, 275), meta=np.ndarray>
- long_name :
- latitude of grid cell center
- units :
- degrees_north
- bounds :
- yv
Array Chunk Bytes 451.00 kB 451.00 kB Shape (205, 275) (205, 275) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray
- Tair(time, y, x)float64dask.array<chunksize=(12, 205, 275), meta=np.ndarray>
- units :
- C
- long_name :
- Surface air temperature
- type_preferred :
- double
- time_rep :
- instantaneous
Array Chunk Bytes 16.24 MB 5.41 MB Shape (36, 205, 275) (12, 205, 275) Count 4 Tasks 3 Chunks Type float64 numpy.ndarray
- title :
- /workspace/jhamman/processed/R1002RBRxaaa01a/lnd/temp/R1002RBRxaaa01a.vic.ha.1979-09-01.nc
- institution :
- U.W.
- source :
- RACM R1002RBRxaaa01a
- output_frequency :
- daily
- output_mode :
- averaged
- convention :
- CF-1.4
- references :
- Based on the initial model of Liang et al., 1994, JGR, 99, 14,415- 14,429.
- comment :
- Output from the Variable Infiltration Capacity (VIC) model.
- nco_openmp_thread_number :
- 1
- NCO :
- "4.6.0"
- history :
- Tue Dec 27 14:15:22 2016: ncatted -a dimensions,,d,, rasm.nc rasm.nc Tue Dec 27 13:38:40 2016: ncks -3 rasm.nc rasm.nc history deleted for brevity
ds.Tair.data.visualize()
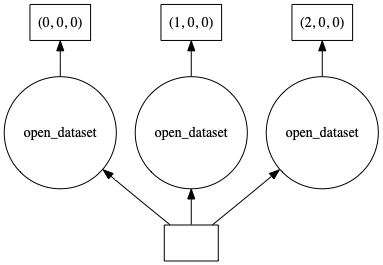
Perform a computation on input dataset#
As you can see, our input dataset is chunked along the time dimension. This produces an xarray dataset with 3 chunks. For illustrative purposes, let’s apply some arbitray computation on our dataset:
result = np.sqrt(np.sin(ds) ** 2 + np.cos(ds) ** 2)
result.Tair.data.visualize()
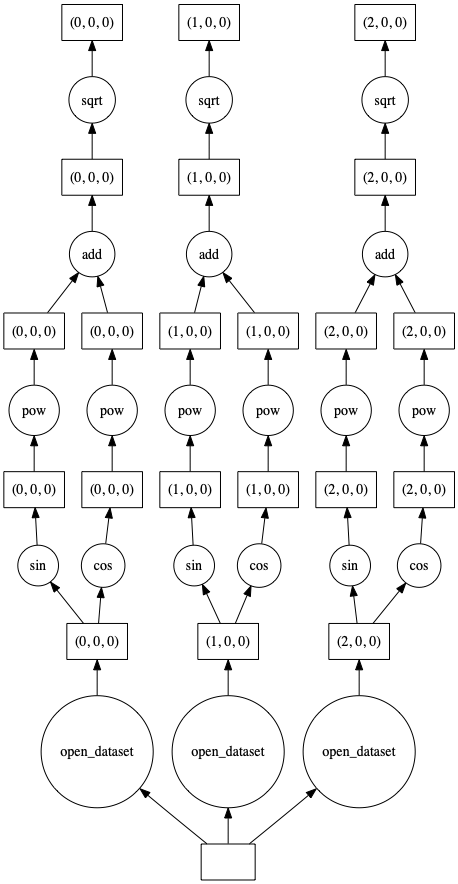
Our resulting xarray dataset has the same dimensions and the same number of chunks as our input dataset.
Create a helper function to split a dataset into sub-datasets#
Our main objective is to save this resulting dataset into multiple netCDF files by saving each chunk in its own netCDF file. To accomplish this, we need to create a function that allows us to split a dataset into sub-datasets for each chunk. The following code snippet was adapted from this comment by Stephan Hoyer.
import itertools
def split_by_chunks(dataset):
chunk_slices = {}
for dim, chunks in dataset.chunks.items():
slices = []
start = 0
for chunk in chunks:
if start >= dataset.sizes[dim]:
break
stop = start + chunk
slices.append(slice(start, stop))
start = stop
chunk_slices[dim] = slices
for slices in itertools.product(*chunk_slices.values()):
selection = dict(zip(chunk_slices.keys(), slices))
yield dataset[selection]
datasets = list(split_by_chunks(result))
Let’s confirm that we have three items corresponding to our three chunks:
print(len(datasets))
3
Each item in the returned list consists of an xarray dataset corresponding to each sliced chunk. Now that we have a list of datasets, we need another helper function that will generate a filepath for us when given an xarray dataset as input:
Create a helper function for generating a filepath#
def create_filepath(ds, prefix='filename', root_path="."):
"""
Generate a filepath when given an xarray dataset
"""
start = ds.time.data[0].strftime("%Y-%m-%d")
end = ds.time.data[-1].strftime("%Y-%m-%d")
filepath = f'{root_path}/{prefix}_{start}_{end}.nc'
return filepath
Let’s run a small test to make sure this function is working as expected:
create_filepath(datasets[1])
'./filename_1981-09-16_1982-08-17.nc'
Let’s create a list of paths to which to save each corresponding dataset.
paths = [create_filepath(ds) for ds in datasets]
paths
['./filename_1980-09-16_1981-08-17.nc',
'./filename_1981-09-16_1982-08-17.nc',
'./filename_1982-09-16_1983-08-17.nc']
Invoke xr.save_mfdataset()#
At this point we have two lists:
datasets: List of datasets to save.paths: List of paths to which to save each corresponding dataset.
We are ready to invoke the xr.save_mfdataset():
xr.save_mfdataset(datasets=datasets, paths=paths)
Confirm that the output files were properly written#
new_ds = xr.open_mfdataset(paths, combine='by_coords')
new_ds
- time: 36
- x: 275
- y: 205
- xc(y, x)float64dask.array<chunksize=(205, 275), meta=np.ndarray>
- long_name :
- longitude of grid cell center
- units :
- degrees_east
- bounds :
- xv
Array Chunk Bytes 451.00 kB 451.00 kB Shape (205, 275) (205, 275) Count 10 Tasks 1 Chunks Type float64 numpy.ndarray - yc(y, x)float64dask.array<chunksize=(205, 275), meta=np.ndarray>
- long_name :
- latitude of grid cell center
- units :
- degrees_north
- bounds :
- yv
Array Chunk Bytes 451.00 kB 451.00 kB Shape (205, 275) (205, 275) Count 10 Tasks 1 Chunks Type float64 numpy.ndarray - time(time)object1980-09-16 12:00:00 ... 1983-08-17 00:00:00
- long_name :
- time
- type_preferred :
- int
array([cftime.DatetimeNoLeap(1980-09-16 12:00:00), cftime.DatetimeNoLeap(1980-10-17 00:00:00), cftime.DatetimeNoLeap(1980-11-16 12:00:00), cftime.DatetimeNoLeap(1980-12-17 00:00:00), cftime.DatetimeNoLeap(1981-01-17 00:00:00), cftime.DatetimeNoLeap(1981-02-15 12:00:00), cftime.DatetimeNoLeap(1981-03-17 00:00:00), cftime.DatetimeNoLeap(1981-04-16 12:00:00), cftime.DatetimeNoLeap(1981-05-17 00:00:00), cftime.DatetimeNoLeap(1981-06-16 12:00:00), cftime.DatetimeNoLeap(1981-07-17 00:00:00), cftime.DatetimeNoLeap(1981-08-17 00:00:00), cftime.DatetimeNoLeap(1981-09-16 12:00:00), cftime.DatetimeNoLeap(1981-10-17 00:00:00), cftime.DatetimeNoLeap(1981-11-16 12:00:00), cftime.DatetimeNoLeap(1981-12-17 00:00:00), cftime.DatetimeNoLeap(1982-01-17 00:00:00), cftime.DatetimeNoLeap(1982-02-15 12:00:00), cftime.DatetimeNoLeap(1982-03-17 00:00:00), cftime.DatetimeNoLeap(1982-04-16 12:00:00), cftime.DatetimeNoLeap(1982-05-17 00:00:00), cftime.DatetimeNoLeap(1982-06-16 12:00:00), cftime.DatetimeNoLeap(1982-07-17 00:00:00), cftime.DatetimeNoLeap(1982-08-17 00:00:00), cftime.DatetimeNoLeap(1982-09-16 12:00:00), cftime.DatetimeNoLeap(1982-10-17 00:00:00), cftime.DatetimeNoLeap(1982-11-16 12:00:00), cftime.DatetimeNoLeap(1982-12-17 00:00:00), cftime.DatetimeNoLeap(1983-01-17 00:00:00), cftime.DatetimeNoLeap(1983-02-15 12:00:00), cftime.DatetimeNoLeap(1983-03-17 00:00:00), cftime.DatetimeNoLeap(1983-04-16 12:00:00), cftime.DatetimeNoLeap(1983-05-17 00:00:00), cftime.DatetimeNoLeap(1983-06-16 12:00:00), cftime.DatetimeNoLeap(1983-07-17 00:00:00), cftime.DatetimeNoLeap(1983-08-17 00:00:00)], dtype=object)
- Tair(time, y, x)float64dask.array<chunksize=(12, 205, 275), meta=np.ndarray>
Array Chunk Bytes 16.24 MB 5.41 MB Shape (36, 205, 275) (12, 205, 275) Count 9 Tasks 3 Chunks Type float64 numpy.ndarray
try:
xr.testing.assert_identical(result, new_ds)
except AssertionError:
print('The datasets are not identical!')
else:
print('The datasets are identical!')
The datasets are identical!
There you have it 🎉🎉🎉🎉
Conclusion#
This article was inspired by a problem that my colleague was running into. I thought it would be useful to document this fix in a blog post. Many thanks to Deepak Cherian for pointing me to some relevant xarray issues and to the @NCAR/xdev team for their valuable and critical feedback on drafts of this article along the way.
