Regridding using xESMF and an existing weights file#
A fairly common request is to use an existing ESMF weights file to regrid a Xarray Dataset (1, 2). Applying weights in general should be easy: read weights then apply them using dot or tensordot on the input dataset.
In the Xarray/Dask/Pangeo ecosystem, xESMF provides an interface to ESMF for convenient regridding, includiing parallelization with Dask. Here we demonstrate how to use an existing ESMF weights file with xESMF specifically for CAM-SE.
CAM-SE is the Community Atmosphere Model with the spectral element dynamical core (Dennis et al, 2011)
The spectral element dynamical core, CAM-SE, is an unstructured grid that supports uniform resolutions based on the equiangular gnomonic cubed-sphere grid as well as a mesh refinement capability with local increases in resolution through conformal mesh refinement. CAM-SE is the default resolution for CESM2 high resolution capabilities … (Lauritzen et al. 2018).
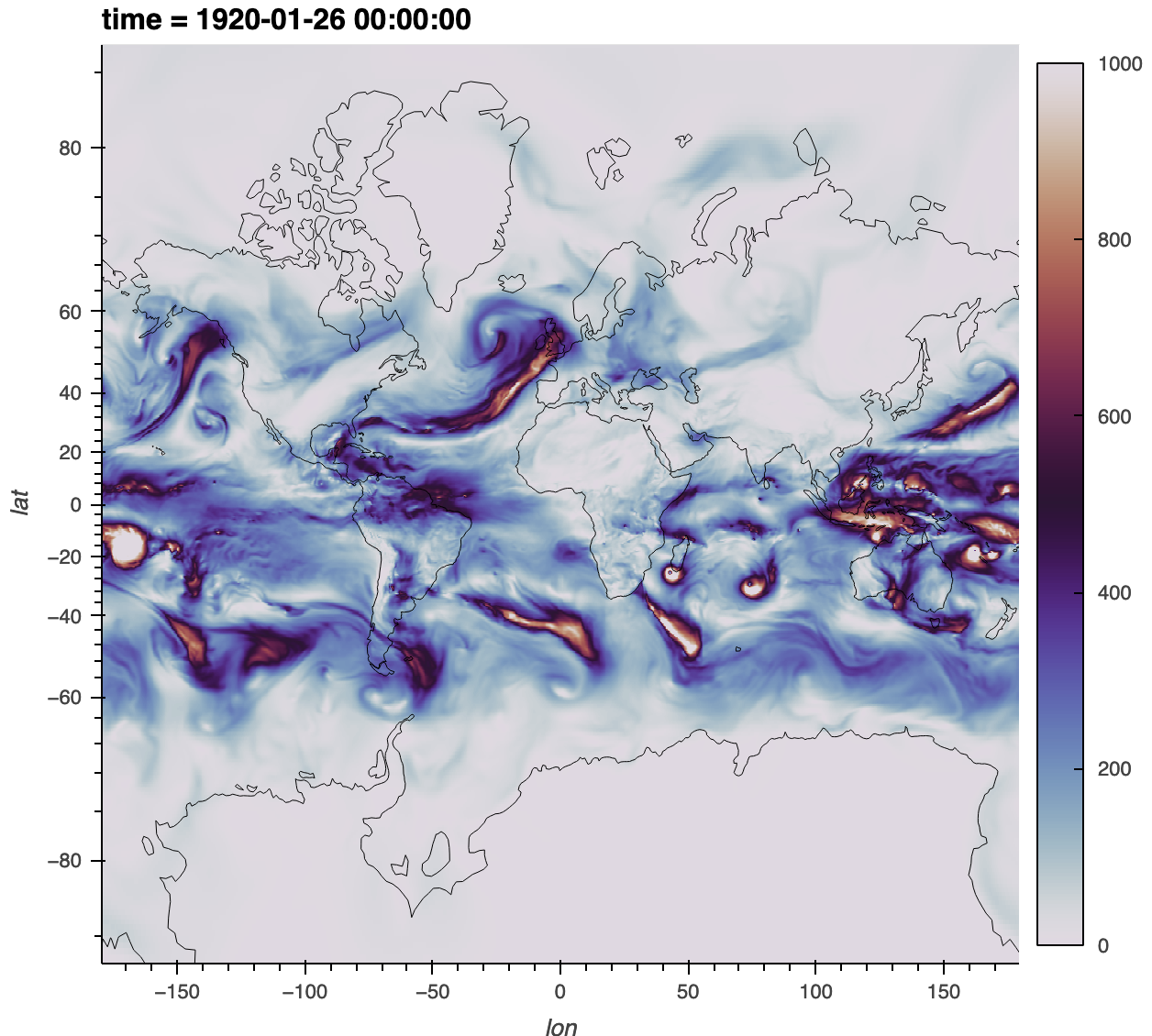
The main challenge is the input dataset has one spatial dimension (ncol), while xESMF is hardcoded to expect two spatial dimensions (lat, lon). We solve that by adding a dummy dimension. At the end, we’ll make this plot of the vertically integrated water vapour transport (IVT)
%load_ext watermark
import hvplot.xarray
import matplotlib.pyplot as plt
import numpy as np
import xarray as xr
import xesmf
%watermark -iv
sys : 3.10.8 | packaged by conda-forge | (main, Nov 22 2022, 08:26:04) [GCC 10.4.0]
numpy : 1.23.5
matplotlib: 3.6.2
xesmf : 0.6.3
json : 2.0.9
xarray : 2022.12.0
hvplot : 0.8.2
Read data#
First some file paths
data_file = "/glade/campaign/collections/cmip/CMIP6/iHESP/BHIST/HR/b.e13.BHISTC5.ne120_t12.cesm-ihesp-hires1.0.30-1920-2100.002/atm/proc/tseries/hour_6I/b.e13.BHISTC5.ne120_t12.cesm-ihesp-hires1.0.30-1920-2100.002.cam.h2.IVT.192001-192912.nc"
weight_file = "/glade/work/shields/SE_grid/map_ne120_to_0.23x0.31_bilinear.nc"
We read in the input data
data_in = xr.open_dataset(data_file, chunks={"time": 50})
data_in
<xarray.Dataset>
Dimensions: (lev: 30, ilev: 31, cosp_prs: 7, nbnd: 2, cosp_tau: 7,
cosp_scol: 50, cosp_ht: 40, cosp_sr: 15, cosp_sza: 5,
ncol: 777602, time: 14600)
Coordinates:
* lev (lev) float64 3.643 7.595 14.36 24.61 ... 957.5 976.3 992.6
* ilev (ilev) float64 2.255 5.032 10.16 18.56 ... 967.5 985.1 1e+03
* cosp_prs (cosp_prs) float64 900.0 740.0 620.0 500.0 375.0 245.0 90.0
* cosp_tau (cosp_tau) float64 0.15 0.8 2.45 6.5 16.2 41.5 219.5
* cosp_scol (cosp_scol) int32 1 2 3 4 5 6 7 8 ... 43 44 45 46 47 48 49 50
* cosp_ht (cosp_ht) float64 240.0 720.0 1.2e+03 ... 1.848e+04 1.896e+04
* cosp_sr (cosp_sr) float64 0.605 2.1 4.0 6.0 ... 70.0 539.5 1.004e+03
* cosp_sza (cosp_sza) float64 0.0 15.0 30.0 45.0 60.0
* time (time) object 1920-01-01 00:00:00 ... 1929-12-31 18:00:00
Dimensions without coordinates: nbnd, ncol
Data variables: (12/35)
hyam (lev) float64 dask.array<chunksize=(30,), meta=np.ndarray>
hybm (lev) float64 dask.array<chunksize=(30,), meta=np.ndarray>
P0 float64 ...
hyai (ilev) float64 dask.array<chunksize=(31,), meta=np.ndarray>
hybi (ilev) float64 dask.array<chunksize=(31,), meta=np.ndarray>
cosp_prs_bnds (cosp_prs, nbnd) float64 dask.array<chunksize=(7, 2), meta=np.ndarray>
... ...
n2ovmr (time) float64 dask.array<chunksize=(50,), meta=np.ndarray>
f11vmr (time) float64 dask.array<chunksize=(50,), meta=np.ndarray>
f12vmr (time) float64 dask.array<chunksize=(50,), meta=np.ndarray>
sol_tsi (time) float64 dask.array<chunksize=(50,), meta=np.ndarray>
nsteph (time) int32 dask.array<chunksize=(50,), meta=np.ndarray>
IVT (time, ncol) float32 dask.array<chunksize=(50, 777602), meta=np.ndarray>
Attributes:
np: 4
ne: 120
Conventions: CF-1.0
source: CAM
case: b.e13.BHISTC5.ne120_t12.cesm-ihesp-hires1.0.30-1920-210...
title: UNSET
logname: nanr
host: login1.frontera.
Version: $Name$
revision_Id: $Id$
initial_file: B.E.13.BHISTC5.ne120_t12.sehires38.003.sunway.cam.i.192...
topography_file: /work/02503/edwardsj/CESM/inputdata//atm/cam/topo/USGS-...- lev: 30
- ilev: 31
- cosp_prs: 7
- nbnd: 2
- cosp_tau: 7
- cosp_scol: 50
- cosp_ht: 40
- cosp_sr: 15
- cosp_sza: 5
- ncol: 777602
- time: 14600
- lev(lev)float643.643 7.595 14.36 ... 976.3 992.6
- long_name :
- hybrid level at midpoints (1000*(A+B))
- units :
- hPa
- positive :
- down
- standard_name :
- atmosphere_hybrid_sigma_pressure_coordinate
- formula_terms :
- a: hyam b: hybm p0: P0 ps: PS
array([ 3.643466, 7.59482 , 14.356632, 24.61222 , 38.2683 , 54.59548 , 72.012451, 87.82123 , 103.317127, 121.547241, 142.994039, 168.22508 , 197.908087, 232.828619, 273.910817, 322.241902, 379.100904, 445.992574, 524.687175, 609.778695, 691.38943 , 763.404481, 820.858369, 859.534767, 887.020249, 912.644547, 936.198398, 957.48548 , 976.325407, 992.556095]) - ilev(ilev)float642.255 5.032 10.16 ... 985.1 1e+03
- long_name :
- hybrid level at interfaces (1000*(A+B))
- units :
- hPa
- positive :
- down
- standard_name :
- atmosphere_hybrid_sigma_pressure_coordinate
- formula_terms :
- a: hyai b: hybi p0: P0 ps: PS
array([ 2.25524 , 5.031692, 10.157947, 18.555317, 30.669123, 45.867477, 63.323483, 80.701418, 94.941042, 111.693211, 131.401271, 154.586807, 181.863353, 213.952821, 251.704417, 296.117216, 348.366588, 409.835219, 482.149929, 567.224421, 652.332969, 730.445892, 796.363071, 845.353667, 873.715866, 900.324631, 924.964462, 947.432335, 967.538625, 985.11219 , 1000. ]) - cosp_prs(cosp_prs)float64900.0 740.0 620.0 ... 245.0 90.0
- long_name :
- COSP Mean ISCCP pressure
- units :
- hPa
- bounds :
- cosp_prs_bnds
array([900., 740., 620., 500., 375., 245., 90.])
- cosp_tau(cosp_tau)float640.15 0.8 2.45 6.5 16.2 41.5 219.5
- long_name :
- COSP Mean ISCCP optical depth
- units :
- 1
- bounds :
- cosp_tau_bnds
array([1.500e-01, 8.000e-01, 2.450e+00, 6.500e+00, 1.620e+01, 4.150e+01, 2.195e+02]) - cosp_scol(cosp_scol)int321 2 3 4 5 6 7 ... 45 46 47 48 49 50
- long_name :
- COSP subcolumn
array([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50], dtype=int32) - cosp_ht(cosp_ht)float64240.0 720.0 ... 1.848e+04 1.896e+04
- long_name :
- COSP Mean Height for lidar and radar simulator outputs
- units :
- m
- bounds :
- cosp_ht_bnds
array([ 240., 720., 1200., 1680., 2160., 2640., 3120., 3600., 4080., 4560., 5040., 5520., 6000., 6480., 6960., 7440., 7920., 8400., 8880., 9360., 9840., 10320., 10800., 11280., 11760., 12240., 12720., 13200., 13680., 14160., 14640., 15120., 15600., 16080., 16560., 17040., 17520., 18000., 18480., 18960.]) - cosp_sr(cosp_sr)float640.605 2.1 4.0 ... 539.5 1.004e+03
- long_name :
- COSP Mean Scattering Ratio for lidar simulator CFAD output
- units :
- 1
- bounds :
- cosp_sr_bnds
array([6.050e-01, 2.100e+00, 4.000e+00, 6.000e+00, 8.500e+00, 1.250e+01, 1.750e+01, 2.250e+01, 2.750e+01, 3.500e+01, 4.500e+01, 5.500e+01, 7.000e+01, 5.395e+02, 1.004e+03]) - cosp_sza(cosp_sza)float640.0 15.0 30.0 45.0 60.0
- long_name :
- COSP Parasol SZA
- units :
- degrees
array([ 0., 15., 30., 45., 60.])
- time(time)object1920-01-01 00:00:00 ... 1929-12-...
- long_name :
- time
- bounds :
- time_bnds
array([cftime.DatetimeNoLeap(1920, 1, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1920, 1, 1, 6, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1920, 1, 1, 12, 0, 0, 0, has_year_zero=True), ..., cftime.DatetimeNoLeap(1929, 12, 31, 6, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1929, 12, 31, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1929, 12, 31, 18, 0, 0, 0, has_year_zero=True)], dtype=object)
- hyam(lev)float64dask.array<chunksize=(30,), meta=np.ndarray>
- long_name :
- hybrid A coefficient at layer midpoints
Array Chunk Bytes 240 B 240 B Shape (30,) (30,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - hybm(lev)float64dask.array<chunksize=(30,), meta=np.ndarray>
- long_name :
- hybrid B coefficient at layer midpoints
Array Chunk Bytes 240 B 240 B Shape (30,) (30,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - P0()float64...
- long_name :
- reference pressure
- units :
- Pa
[1 values with dtype=float64]
- hyai(ilev)float64dask.array<chunksize=(31,), meta=np.ndarray>
- long_name :
- hybrid A coefficient at layer interfaces
Array Chunk Bytes 248 B 248 B Shape (31,) (31,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - hybi(ilev)float64dask.array<chunksize=(31,), meta=np.ndarray>
- long_name :
- hybrid B coefficient at layer interfaces
Array Chunk Bytes 248 B 248 B Shape (31,) (31,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - cosp_prs_bnds(cosp_prs, nbnd)float64dask.array<chunksize=(7, 2), meta=np.ndarray>
Array Chunk Bytes 112 B 112 B Shape (7, 2) (7, 2) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - cosp_tau_bnds(cosp_tau, nbnd)float64dask.array<chunksize=(7, 2), meta=np.ndarray>
Array Chunk Bytes 112 B 112 B Shape (7, 2) (7, 2) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - cosp_ht_bnds(cosp_ht, nbnd)float64dask.array<chunksize=(40, 2), meta=np.ndarray>
Array Chunk Bytes 640 B 640 B Shape (40, 2) (40, 2) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - cosp_sr_bnds(cosp_sr, nbnd)float64dask.array<chunksize=(15, 2), meta=np.ndarray>
Array Chunk Bytes 240 B 240 B Shape (15, 2) (15, 2) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - lat(ncol)float64dask.array<chunksize=(777602,), meta=np.ndarray>
- long_name :
- latitude
- units :
- degrees_north
Array Chunk Bytes 5.93 MiB 5.93 MiB Shape (777602,) (777602,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - area(ncol)float64dask.array<chunksize=(777602,), meta=np.ndarray>
Array Chunk Bytes 5.93 MiB 5.93 MiB Shape (777602,) (777602,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - lon(ncol)float64dask.array<chunksize=(777602,), meta=np.ndarray>
- long_name :
- longitude
- units :
- degrees_east
Array Chunk Bytes 5.93 MiB 5.93 MiB Shape (777602,) (777602,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - ntrm()int32...
- long_name :
- spectral truncation parameter M
[1 values with dtype=int32]
- ntrn()int32...
- long_name :
- spectral truncation parameter N
[1 values with dtype=int32]
- ntrk()int32...
- long_name :
- spectral truncation parameter K
[1 values with dtype=int32]
- ndbase()int32...
- long_name :
- base day
[1 values with dtype=int32]
- nsbase()int32...
- long_name :
- seconds of base day
[1 values with dtype=int32]
- nbdate()int32...
- long_name :
- base date (YYYYMMDD)
[1 values with dtype=int32]
- nbsec()int32...
- long_name :
- seconds of base date
[1 values with dtype=int32]
- mdt()int32...
- long_name :
- timestep
- units :
- s
[1 values with dtype=int32]
- date(time)int32dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- current date (YYYYMMDD)
Array Chunk Bytes 57.03 kiB 200 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type int32 numpy.ndarray - datesec(time)int32dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- current seconds of current date
Array Chunk Bytes 57.03 kiB 200 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type int32 numpy.ndarray - time_bnds(time, nbnd)objectdask.array<chunksize=(50, 2), meta=np.ndarray>
- long_name :
- time interval endpoints
Array Chunk Bytes 228.12 kiB 800 B Shape (14600, 2) (50, 2) Dask graph 292 chunks in 2 graph layers Data type object numpy.ndarray - date_written(time)|S8dask.array<chunksize=(50,), meta=np.ndarray>
Array Chunk Bytes 114.06 kiB 400 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type |S8 numpy.ndarray - time_written(time)|S8dask.array<chunksize=(50,), meta=np.ndarray>
Array Chunk Bytes 114.06 kiB 400 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type |S8 numpy.ndarray - ndcur(time)int32dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- current day (from base day)
Array Chunk Bytes 57.03 kiB 200 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type int32 numpy.ndarray - nscur(time)int32dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- current seconds of current day
Array Chunk Bytes 57.03 kiB 200 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type int32 numpy.ndarray - co2vmr(time)float64dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- co2 volume mixing ratio
Array Chunk Bytes 114.06 kiB 400 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type float64 numpy.ndarray - ch4vmr(time)float64dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- ch4 volume mixing ratio
Array Chunk Bytes 114.06 kiB 400 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type float64 numpy.ndarray - n2ovmr(time)float64dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- n2o volume mixing ratio
Array Chunk Bytes 114.06 kiB 400 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type float64 numpy.ndarray - f11vmr(time)float64dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- f11 volume mixing ratio
Array Chunk Bytes 114.06 kiB 400 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type float64 numpy.ndarray - f12vmr(time)float64dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- f12 volume mixing ratio
Array Chunk Bytes 114.06 kiB 400 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type float64 numpy.ndarray - sol_tsi(time)float64dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- total solar irradiance
- units :
- W/m2
Array Chunk Bytes 114.06 kiB 400 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type float64 numpy.ndarray - nsteph(time)int32dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- current timestep
Array Chunk Bytes 57.03 kiB 200 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type int32 numpy.ndarray - IVT(time, ncol)float32dask.array<chunksize=(50, 777602), meta=np.ndarray>
- units :
- kg/m/s
- long_name :
- Total (vertically integrated) vapor transport
Array Chunk Bytes 42.29 GiB 148.32 MiB Shape (14600, 777602) (50, 777602) Dask graph 292 chunks in 2 graph layers Data type float32 numpy.ndarray
- levPandasIndex
PandasIndex(Float64Index([ 3.64346569404006, 7.594819646328688, 14.356632251292467, 24.612220004200935, 38.26829977333546, 54.59547974169254, 72.01245054602623, 87.82123029232025, 103.31712663173676, 121.54724076390266, 142.99403876066208, 168.22507977485657, 197.9080867022276, 232.82861895859241, 273.9108167588711, 322.2419023513794, 379.10090386867523, 445.992574095726, 524.6871747076511, 609.7786948084831, 691.3894303143024, 763.404481112957, 820.8583686500788, 859.5347665250301, 887.0202489197254, 912.644546944648, 936.1983984708786, 957.485479535535, 976.325407391414, 992.556095123291], dtype='float64', name='lev')) - ilevPandasIndex
PandasIndex(Float64Index([ 2.255239523947239, 5.031691864132881, 10.15794742852449, 18.55531707406044, 30.66912293434143, 45.86747661232948, 63.3234828710556, 80.70141822099686, 94.94104236364365, 111.69321089982991, 131.4012706279755, 154.5868068933487, 181.8633526563644, 213.95282074809072, 251.70441716909409, 296.11721634864807, 348.3665883541107, 409.8352193832398, 482.14992880821234, 567.22442060709, 652.3329690098763, 730.4458916187286, 796.3630706071854, 845.3536666929722, 873.7158663570881, 900.3246314823627, 924.9644624069333, 947.4323345348239, 967.5386245362461, 985.112190246582, 1000.0], dtype='float64', name='ilev')) - cosp_prsPandasIndex
PandasIndex(Float64Index([900.0, 740.0, 620.0, 500.0, 375.0, 245.0, 90.0], dtype='float64', name='cosp_prs'))
- cosp_tauPandasIndex
PandasIndex(Float64Index([0.15, 0.8, 2.45, 6.5, 16.2, 41.5, 219.5], dtype='float64', name='cosp_tau'))
- cosp_scolPandasIndex
PandasIndex(Int64Index([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50], dtype='int64', name='cosp_scol')) - cosp_htPandasIndex
PandasIndex(Float64Index([ 240.0, 720.0, 1200.0, 1680.0, 2160.0, 2640.0, 3120.0, 3600.0, 4080.0, 4560.0, 5040.0, 5520.0, 6000.0, 6480.0, 6960.0, 7440.0, 7920.0, 8400.0, 8880.0, 9360.0, 9840.0, 10320.0, 10800.0, 11280.0, 11760.0, 12240.0, 12720.0, 13200.0, 13680.0, 14160.0, 14640.0, 15120.0, 15600.0, 16080.0, 16560.0, 17040.0, 17520.0, 18000.0, 18480.0, 18960.0], dtype='float64', name='cosp_ht')) - cosp_srPandasIndex
PandasIndex(Float64Index([ 0.605, 2.1, 4.0, 6.0, 8.5, 12.5, 17.5, 22.5, 27.5, 35.0, 45.0, 55.0, 70.0, 539.5, 1004.0], dtype='float64', name='cosp_sr')) - cosp_szaPandasIndex
PandasIndex(Float64Index([0.0, 15.0, 30.0, 45.0, 60.0], dtype='float64', name='cosp_sza'))
- timePandasIndex
PandasIndex(CFTimeIndex([1920-01-01 00:00:00, 1920-01-01 06:00:00, 1920-01-01 12:00:00, 1920-01-01 18:00:00, 1920-01-02 00:00:00, 1920-01-02 06:00:00, 1920-01-02 12:00:00, 1920-01-02 18:00:00, 1920-01-03 00:00:00, 1920-01-03 06:00:00, ... 1929-12-29 12:00:00, 1929-12-29 18:00:00, 1929-12-30 00:00:00, 1929-12-30 06:00:00, 1929-12-30 12:00:00, 1929-12-30 18:00:00, 1929-12-31 00:00:00, 1929-12-31 06:00:00, 1929-12-31 12:00:00, 1929-12-31 18:00:00], dtype='object', length=14600, calendar='noleap', freq='6H'))
- np :
- 4
- ne :
- 120
- Conventions :
- CF-1.0
- source :
- CAM
- case :
- b.e13.BHISTC5.ne120_t12.cesm-ihesp-hires1.0.30-1920-2100.002
- title :
- UNSET
- logname :
- nanr
- host :
- login1.frontera.
- Version :
- $Name$
- revision_Id :
- $Id$
- initial_file :
- B.E.13.BHISTC5.ne120_t12.sehires38.003.sunway.cam.i.1920-01-01-00000.nc
- topography_file :
- /work/02503/edwardsj/CESM/inputdata//atm/cam/topo/USGS-gtopo30_ne120np4_16xdel2-PFC-consistentSGH.nc
Here’s the primary data variable IVT for integrated vapour transport with one spatial dimension: ncol
data_in.IVT
<xarray.DataArray 'IVT' (time: 14600, ncol: 777602)>
dask.array<open_dataset-124517cf72a30883d5a3c70220985aeeIVT, shape=(14600, 777602), dtype=float32, chunksize=(50, 777602), chunktype=numpy.ndarray>
Coordinates:
* time (time) object 1920-01-01 00:00:00 ... 1929-12-31 18:00:00
Dimensions without coordinates: ncol
Attributes:
units: kg/m/s
long_name: Total (vertically integrated) vapor transport- time: 14600
- ncol: 777602
- dask.array<chunksize=(50, 777602), meta=np.ndarray>
Array Chunk Bytes 42.29 GiB 148.32 MiB Shape (14600, 777602) (50, 777602) Dask graph 292 chunks in 2 graph layers Data type float32 numpy.ndarray - time(time)object1920-01-01 00:00:00 ... 1929-12-...
- long_name :
- time
- bounds :
- time_bnds
array([cftime.DatetimeNoLeap(1920, 1, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1920, 1, 1, 6, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1920, 1, 1, 12, 0, 0, 0, has_year_zero=True), ..., cftime.DatetimeNoLeap(1929, 12, 31, 6, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1929, 12, 31, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1929, 12, 31, 18, 0, 0, 0, has_year_zero=True)], dtype=object)
- timePandasIndex
PandasIndex(CFTimeIndex([1920-01-01 00:00:00, 1920-01-01 06:00:00, 1920-01-01 12:00:00, 1920-01-01 18:00:00, 1920-01-02 00:00:00, 1920-01-02 06:00:00, 1920-01-02 12:00:00, 1920-01-02 18:00:00, 1920-01-03 00:00:00, 1920-01-03 06:00:00, ... 1929-12-29 12:00:00, 1929-12-29 18:00:00, 1929-12-30 00:00:00, 1929-12-30 06:00:00, 1929-12-30 12:00:00, 1929-12-30 18:00:00, 1929-12-31 00:00:00, 1929-12-31 06:00:00, 1929-12-31 12:00:00, 1929-12-31 18:00:00], dtype='object', length=14600, calendar='noleap', freq='6H'))
- units :
- kg/m/s
- long_name :
- Total (vertically integrated) vapor transport
And here’s what an ESMF weights file looks like:
weights = xr.open_dataset(weight_file)
weights
<xarray.Dataset>
Dimensions: (src_grid_rank: 1, dst_grid_rank: 2, n_a: 777602,
n_b: 884736, nv_a: 3, nv_b: 4, n_s: 2654208)
Dimensions without coordinates: src_grid_rank, dst_grid_rank, n_a, n_b, nv_a,
nv_b, n_s
Data variables: (12/19)
src_grid_dims (src_grid_rank) int32 ...
dst_grid_dims (dst_grid_rank) int32 ...
yc_a (n_a) float64 ...
yc_b (n_b) float64 ...
xc_a (n_a) float64 ...
xc_b (n_b) float64 ...
... ...
area_b (n_b) float64 ...
frac_a (n_a) float64 ...
frac_b (n_b) float64 ...
col (n_s) int32 ...
row (n_s) int32 ...
S (n_s) float64 ...
Attributes:
title: ESMF Offline Regridding Weight Generator
normalization: destarea
map_method: Bilinear remapping
ESMF_regrid_method: Bilinear
conventions: NCAR-CSM
domain_a: /glade/scratch/shields/regridded/mapfiles/ne120.nc
domain_b: /glade/scratch/shields/regridded/mapfiles/0.23x0.31.nc
grid_file_src: /glade/scratch/shields/regridded/mapfiles/ne120.nc
grid_file_dst: /glade/scratch/shields/regridded/mapfiles/0.23x0.31.nc
CVS_revision: 7.1.0r- src_grid_rank: 1
- dst_grid_rank: 2
- n_a: 777602
- n_b: 884736
- nv_a: 3
- nv_b: 4
- n_s: 2654208
- src_grid_dims(src_grid_rank)int32...
[1 values with dtype=int32]
- dst_grid_dims(dst_grid_rank)int32...
[2 values with dtype=int32]
- yc_a(n_a)float64...
- units :
- degrees
[777602 values with dtype=float64]
- yc_b(n_b)float64...
- units :
- degrees
[884736 values with dtype=float64]
- xc_a(n_a)float64...
- units :
- degrees
[777602 values with dtype=float64]
- xc_b(n_b)float64...
- units :
- degrees
[884736 values with dtype=float64]
- yv_a(n_a, nv_a)float64...
- units :
- degrees
[2332806 values with dtype=float64]
- xv_a(n_a, nv_a)float64...
- units :
- degrees
[2332806 values with dtype=float64]
- yv_b(n_b, nv_b)float64...
- units :
- degrees
[3538944 values with dtype=float64]
- xv_b(n_b, nv_b)float64...
- units :
- degrees
[3538944 values with dtype=float64]
- mask_a(n_a)int32...
- units :
- unitless
[777602 values with dtype=int32]
- mask_b(n_b)int32...
- units :
- unitless
[884736 values with dtype=int32]
- area_a(n_a)float64...
- units :
- square radians
[777602 values with dtype=float64]
- area_b(n_b)float64...
- units :
- square radians
[884736 values with dtype=float64]
- frac_a(n_a)float64...
- units :
- unitless
[777602 values with dtype=float64]
- frac_b(n_b)float64...
- units :
- unitless
[884736 values with dtype=float64]
- col(n_s)int32...
[2654208 values with dtype=int32]
- row(n_s)int32...
[2654208 values with dtype=int32]
- S(n_s)float64...
[2654208 values with dtype=float64]
- title :
- ESMF Offline Regridding Weight Generator
- normalization :
- destarea
- map_method :
- Bilinear remapping
- ESMF_regrid_method :
- Bilinear
- conventions :
- NCAR-CSM
- domain_a :
- /glade/scratch/shields/regridded/mapfiles/ne120.nc
- domain_b :
- /glade/scratch/shields/regridded/mapfiles/0.23x0.31.nc
- grid_file_src :
- /glade/scratch/shields/regridded/mapfiles/ne120.nc
- grid_file_dst :
- /glade/scratch/shields/regridded/mapfiles/0.23x0.31.nc
- CVS_revision :
- 7.1.0r
Regridding with xESMF#
Th primary xESMF interface is the Regridder class.
regridder = xesmf.Regridder(
ds_in,
ds_out,
weights=weight_file,
method="bilinear",
reuse_weights=True,
periodic=True,
)
It requires input grid information in ds_in and output grid information in ds_out. Both files need to have variables lat, lon or variables that can be identified as “latitude” and “longitude” using CF metadata.
We can construct this information from the weights file:
Read the weights file#
weights = xr.open_dataset(weight_file)
# input variable shape
in_shape = weights.src_grid_dims.load().data
# Since xESMF expects 2D vars, we'll insert a dummy dimension of size-1
if len(in_shape) == 1:
in_shape = [1, in_shape.item()]
# output variable shape
out_shape = weights.dst_grid_dims.load().data.tolist()[::-1]
in_shape, out_shape
([1, 777602], [768, 1152])
Construct a Regridder.#
First we make dummy input dataset with lat of size 1 and lon of same size as ncol, and an output dataset with the right lat, lon. For the latter, we use xc_b and yc_b as locations of the centers.
Note
We are assuming a rectilinear output grid, but this could be modified for a curvilinear grid. (Question: Is there a way to identify this from the weights file?)
As a reminder, we use lat, lon dimension names because this is hardcoded in to xESMF.
dummy_in = xr.Dataset(
{
"lat": ("lat", np.empty((in_shape[0],))),
"lon": ("lon", np.empty((in_shape[1],))),
}
)
dummy_out = xr.Dataset(
{
"lat": ("lat", weights.yc_b.data.reshape(out_shape)[:, 0]),
"lon": ("lon", weights.xc_b.data.reshape(out_shape)[0, :]),
}
)
regridder = xesmf.Regridder(
dummy_in,
dummy_out,
weights=weight_file,
method="bilinear",
reuse_weights=True,
periodic=True,
)
regridder
xESMF Regridder
Regridding algorithm: bilinear
Weight filename: bilinear_1x777602_768x1152_peri.nc
Reuse pre-computed weights? True
Input grid shape: (1, 777602)
Output grid shape: (768, 1152)
Periodic in longitude? True
This works but note that a lot of metadata here is wrong like “Weight filename”
Apply the Regridder#
Next to apply the regridder, we’ll use DataArray.expand_dims insert a new "dummy" dimension before the ncol dimension using axis=-2. To be safe, we force ncol to be the last dimension using transpose. We do this only for variables that already have the ncol dimension.
Here I’m using the name dummy so that we are clear that it is fake.
vars_with_ncol = [name for name in data_in.variables if "ncol" in data_in[name].dims]
updated = data_in.copy().update(
data_in[vars_with_ncol].transpose(..., "ncol").expand_dims("dummy", axis=-2)
)
updated
<xarray.Dataset>
Dimensions: (lev: 30, ilev: 31, cosp_prs: 7, nbnd: 2, cosp_tau: 7,
cosp_scol: 50, cosp_ht: 40, cosp_sr: 15, cosp_sza: 5,
dummy: 1, ncol: 777602, time: 14600)
Coordinates:
* lev (lev) float64 3.643 7.595 14.36 24.61 ... 957.5 976.3 992.6
* ilev (ilev) float64 2.255 5.032 10.16 18.56 ... 967.5 985.1 1e+03
* cosp_prs (cosp_prs) float64 900.0 740.0 620.0 500.0 375.0 245.0 90.0
* cosp_tau (cosp_tau) float64 0.15 0.8 2.45 6.5 16.2 41.5 219.5
* cosp_scol (cosp_scol) int32 1 2 3 4 5 6 7 8 ... 43 44 45 46 47 48 49 50
* cosp_ht (cosp_ht) float64 240.0 720.0 1.2e+03 ... 1.848e+04 1.896e+04
* cosp_sr (cosp_sr) float64 0.605 2.1 4.0 6.0 ... 70.0 539.5 1.004e+03
* cosp_sza (cosp_sza) float64 0.0 15.0 30.0 45.0 60.0
* time (time) object 1920-01-01 00:00:00 ... 1929-12-31 18:00:00
Dimensions without coordinates: nbnd, dummy, ncol
Data variables: (12/35)
hyam (lev) float64 dask.array<chunksize=(30,), meta=np.ndarray>
hybm (lev) float64 dask.array<chunksize=(30,), meta=np.ndarray>
P0 float64 ...
hyai (ilev) float64 dask.array<chunksize=(31,), meta=np.ndarray>
hybi (ilev) float64 dask.array<chunksize=(31,), meta=np.ndarray>
cosp_prs_bnds (cosp_prs, nbnd) float64 dask.array<chunksize=(7, 2), meta=np.ndarray>
... ...
n2ovmr (time) float64 dask.array<chunksize=(50,), meta=np.ndarray>
f11vmr (time) float64 dask.array<chunksize=(50,), meta=np.ndarray>
f12vmr (time) float64 dask.array<chunksize=(50,), meta=np.ndarray>
sol_tsi (time) float64 dask.array<chunksize=(50,), meta=np.ndarray>
nsteph (time) int32 dask.array<chunksize=(50,), meta=np.ndarray>
IVT (time, dummy, ncol) float32 dask.array<chunksize=(50, 1, 777602), meta=np.ndarray>
Attributes:
np: 4
ne: 120
Conventions: CF-1.0
source: CAM
case: b.e13.BHISTC5.ne120_t12.cesm-ihesp-hires1.0.30-1920-210...
title: UNSET
logname: nanr
host: login1.frontera.
Version: $Name$
revision_Id: $Id$
initial_file: B.E.13.BHISTC5.ne120_t12.sehires38.003.sunway.cam.i.192...
topography_file: /work/02503/edwardsj/CESM/inputdata//atm/cam/topo/USGS-...- lev: 30
- ilev: 31
- cosp_prs: 7
- nbnd: 2
- cosp_tau: 7
- cosp_scol: 50
- cosp_ht: 40
- cosp_sr: 15
- cosp_sza: 5
- dummy: 1
- ncol: 777602
- time: 14600
- lev(lev)float643.643 7.595 14.36 ... 976.3 992.6
- long_name :
- hybrid level at midpoints (1000*(A+B))
- units :
- hPa
- positive :
- down
- standard_name :
- atmosphere_hybrid_sigma_pressure_coordinate
- formula_terms :
- a: hyam b: hybm p0: P0 ps: PS
array([ 3.643466, 7.59482 , 14.356632, 24.61222 , 38.2683 , 54.59548 , 72.012451, 87.82123 , 103.317127, 121.547241, 142.994039, 168.22508 , 197.908087, 232.828619, 273.910817, 322.241902, 379.100904, 445.992574, 524.687175, 609.778695, 691.38943 , 763.404481, 820.858369, 859.534767, 887.020249, 912.644547, 936.198398, 957.48548 , 976.325407, 992.556095]) - ilev(ilev)float642.255 5.032 10.16 ... 985.1 1e+03
- long_name :
- hybrid level at interfaces (1000*(A+B))
- units :
- hPa
- positive :
- down
- standard_name :
- atmosphere_hybrid_sigma_pressure_coordinate
- formula_terms :
- a: hyai b: hybi p0: P0 ps: PS
array([ 2.25524 , 5.031692, 10.157947, 18.555317, 30.669123, 45.867477, 63.323483, 80.701418, 94.941042, 111.693211, 131.401271, 154.586807, 181.863353, 213.952821, 251.704417, 296.117216, 348.366588, 409.835219, 482.149929, 567.224421, 652.332969, 730.445892, 796.363071, 845.353667, 873.715866, 900.324631, 924.964462, 947.432335, 967.538625, 985.11219 , 1000. ]) - cosp_prs(cosp_prs)float64900.0 740.0 620.0 ... 245.0 90.0
- long_name :
- COSP Mean ISCCP pressure
- units :
- hPa
- bounds :
- cosp_prs_bnds
array([900., 740., 620., 500., 375., 245., 90.])
- cosp_tau(cosp_tau)float640.15 0.8 2.45 6.5 16.2 41.5 219.5
- long_name :
- COSP Mean ISCCP optical depth
- units :
- 1
- bounds :
- cosp_tau_bnds
array([1.500e-01, 8.000e-01, 2.450e+00, 6.500e+00, 1.620e+01, 4.150e+01, 2.195e+02]) - cosp_scol(cosp_scol)int321 2 3 4 5 6 7 ... 45 46 47 48 49 50
- long_name :
- COSP subcolumn
array([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50], dtype=int32) - cosp_ht(cosp_ht)float64240.0 720.0 ... 1.848e+04 1.896e+04
- long_name :
- COSP Mean Height for lidar and radar simulator outputs
- units :
- m
- bounds :
- cosp_ht_bnds
array([ 240., 720., 1200., 1680., 2160., 2640., 3120., 3600., 4080., 4560., 5040., 5520., 6000., 6480., 6960., 7440., 7920., 8400., 8880., 9360., 9840., 10320., 10800., 11280., 11760., 12240., 12720., 13200., 13680., 14160., 14640., 15120., 15600., 16080., 16560., 17040., 17520., 18000., 18480., 18960.]) - cosp_sr(cosp_sr)float640.605 2.1 4.0 ... 539.5 1.004e+03
- long_name :
- COSP Mean Scattering Ratio for lidar simulator CFAD output
- units :
- 1
- bounds :
- cosp_sr_bnds
array([6.050e-01, 2.100e+00, 4.000e+00, 6.000e+00, 8.500e+00, 1.250e+01, 1.750e+01, 2.250e+01, 2.750e+01, 3.500e+01, 4.500e+01, 5.500e+01, 7.000e+01, 5.395e+02, 1.004e+03]) - cosp_sza(cosp_sza)float640.0 15.0 30.0 45.0 60.0
- long_name :
- COSP Parasol SZA
- units :
- degrees
array([ 0., 15., 30., 45., 60.])
- time(time)object1920-01-01 00:00:00 ... 1929-12-...
- long_name :
- time
- bounds :
- time_bnds
array([cftime.DatetimeNoLeap(1920, 1, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1920, 1, 1, 6, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1920, 1, 1, 12, 0, 0, 0, has_year_zero=True), ..., cftime.DatetimeNoLeap(1929, 12, 31, 6, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1929, 12, 31, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1929, 12, 31, 18, 0, 0, 0, has_year_zero=True)], dtype=object)
- hyam(lev)float64dask.array<chunksize=(30,), meta=np.ndarray>
- long_name :
- hybrid A coefficient at layer midpoints
Array Chunk Bytes 240 B 240 B Shape (30,) (30,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - hybm(lev)float64dask.array<chunksize=(30,), meta=np.ndarray>
- long_name :
- hybrid B coefficient at layer midpoints
Array Chunk Bytes 240 B 240 B Shape (30,) (30,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - P0()float64...
- long_name :
- reference pressure
- units :
- Pa
[1 values with dtype=float64]
- hyai(ilev)float64dask.array<chunksize=(31,), meta=np.ndarray>
- long_name :
- hybrid A coefficient at layer interfaces
Array Chunk Bytes 248 B 248 B Shape (31,) (31,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - hybi(ilev)float64dask.array<chunksize=(31,), meta=np.ndarray>
- long_name :
- hybrid B coefficient at layer interfaces
Array Chunk Bytes 248 B 248 B Shape (31,) (31,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - cosp_prs_bnds(cosp_prs, nbnd)float64dask.array<chunksize=(7, 2), meta=np.ndarray>
Array Chunk Bytes 112 B 112 B Shape (7, 2) (7, 2) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - cosp_tau_bnds(cosp_tau, nbnd)float64dask.array<chunksize=(7, 2), meta=np.ndarray>
Array Chunk Bytes 112 B 112 B Shape (7, 2) (7, 2) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - cosp_ht_bnds(cosp_ht, nbnd)float64dask.array<chunksize=(40, 2), meta=np.ndarray>
Array Chunk Bytes 640 B 640 B Shape (40, 2) (40, 2) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - cosp_sr_bnds(cosp_sr, nbnd)float64dask.array<chunksize=(15, 2), meta=np.ndarray>
Array Chunk Bytes 240 B 240 B Shape (15, 2) (15, 2) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - lat(dummy, ncol)float64dask.array<chunksize=(1, 777602), meta=np.ndarray>
- long_name :
- latitude
- units :
- degrees_north
Array Chunk Bytes 5.93 MiB 5.93 MiB Shape (1, 777602) (1, 777602) Dask graph 1 chunks in 3 graph layers Data type float64 numpy.ndarray - area(dummy, ncol)float64dask.array<chunksize=(1, 777602), meta=np.ndarray>
Array Chunk Bytes 5.93 MiB 5.93 MiB Shape (1, 777602) (1, 777602) Dask graph 1 chunks in 3 graph layers Data type float64 numpy.ndarray - lon(dummy, ncol)float64dask.array<chunksize=(1, 777602), meta=np.ndarray>
- long_name :
- longitude
- units :
- degrees_east
Array Chunk Bytes 5.93 MiB 5.93 MiB Shape (1, 777602) (1, 777602) Dask graph 1 chunks in 3 graph layers Data type float64 numpy.ndarray - ntrm()int32...
- long_name :
- spectral truncation parameter M
[1 values with dtype=int32]
- ntrn()int32...
- long_name :
- spectral truncation parameter N
[1 values with dtype=int32]
- ntrk()int32...
- long_name :
- spectral truncation parameter K
[1 values with dtype=int32]
- ndbase()int32...
- long_name :
- base day
[1 values with dtype=int32]
- nsbase()int32...
- long_name :
- seconds of base day
[1 values with dtype=int32]
- nbdate()int32...
- long_name :
- base date (YYYYMMDD)
[1 values with dtype=int32]
- nbsec()int32...
- long_name :
- seconds of base date
[1 values with dtype=int32]
- mdt()int32...
- long_name :
- timestep
- units :
- s
[1 values with dtype=int32]
- date(time)int32dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- current date (YYYYMMDD)
Array Chunk Bytes 57.03 kiB 200 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type int32 numpy.ndarray - datesec(time)int32dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- current seconds of current date
Array Chunk Bytes 57.03 kiB 200 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type int32 numpy.ndarray - time_bnds(time, nbnd)objectdask.array<chunksize=(50, 2), meta=np.ndarray>
- long_name :
- time interval endpoints
Array Chunk Bytes 228.12 kiB 800 B Shape (14600, 2) (50, 2) Dask graph 292 chunks in 2 graph layers Data type object numpy.ndarray - date_written(time)|S8dask.array<chunksize=(50,), meta=np.ndarray>
Array Chunk Bytes 114.06 kiB 400 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type |S8 numpy.ndarray - time_written(time)|S8dask.array<chunksize=(50,), meta=np.ndarray>
Array Chunk Bytes 114.06 kiB 400 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type |S8 numpy.ndarray - ndcur(time)int32dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- current day (from base day)
Array Chunk Bytes 57.03 kiB 200 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type int32 numpy.ndarray - nscur(time)int32dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- current seconds of current day
Array Chunk Bytes 57.03 kiB 200 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type int32 numpy.ndarray - co2vmr(time)float64dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- co2 volume mixing ratio
Array Chunk Bytes 114.06 kiB 400 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type float64 numpy.ndarray - ch4vmr(time)float64dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- ch4 volume mixing ratio
Array Chunk Bytes 114.06 kiB 400 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type float64 numpy.ndarray - n2ovmr(time)float64dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- n2o volume mixing ratio
Array Chunk Bytes 114.06 kiB 400 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type float64 numpy.ndarray - f11vmr(time)float64dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- f11 volume mixing ratio
Array Chunk Bytes 114.06 kiB 400 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type float64 numpy.ndarray - f12vmr(time)float64dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- f12 volume mixing ratio
Array Chunk Bytes 114.06 kiB 400 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type float64 numpy.ndarray - sol_tsi(time)float64dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- total solar irradiance
- units :
- W/m2
Array Chunk Bytes 114.06 kiB 400 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type float64 numpy.ndarray - nsteph(time)int32dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- current timestep
Array Chunk Bytes 57.03 kiB 200 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type int32 numpy.ndarray - IVT(time, dummy, ncol)float32dask.array<chunksize=(50, 1, 777602), meta=np.ndarray>
- units :
- kg/m/s
- long_name :
- Total (vertically integrated) vapor transport
Array Chunk Bytes 42.29 GiB 148.32 MiB Shape (14600, 1, 777602) (50, 1, 777602) Dask graph 292 chunks in 4 graph layers Data type float32 numpy.ndarray
- levPandasIndex
PandasIndex(Float64Index([ 3.64346569404006, 7.594819646328688, 14.356632251292467, 24.612220004200935, 38.26829977333546, 54.59547974169254, 72.01245054602623, 87.82123029232025, 103.31712663173676, 121.54724076390266, 142.99403876066208, 168.22507977485657, 197.9080867022276, 232.82861895859241, 273.9108167588711, 322.2419023513794, 379.10090386867523, 445.992574095726, 524.6871747076511, 609.7786948084831, 691.3894303143024, 763.404481112957, 820.8583686500788, 859.5347665250301, 887.0202489197254, 912.644546944648, 936.1983984708786, 957.485479535535, 976.325407391414, 992.556095123291], dtype='float64', name='lev')) - ilevPandasIndex
PandasIndex(Float64Index([ 2.255239523947239, 5.031691864132881, 10.15794742852449, 18.55531707406044, 30.66912293434143, 45.86747661232948, 63.3234828710556, 80.70141822099686, 94.94104236364365, 111.69321089982991, 131.4012706279755, 154.5868068933487, 181.8633526563644, 213.95282074809072, 251.70441716909409, 296.11721634864807, 348.3665883541107, 409.8352193832398, 482.14992880821234, 567.22442060709, 652.3329690098763, 730.4458916187286, 796.3630706071854, 845.3536666929722, 873.7158663570881, 900.3246314823627, 924.9644624069333, 947.4323345348239, 967.5386245362461, 985.112190246582, 1000.0], dtype='float64', name='ilev')) - cosp_prsPandasIndex
PandasIndex(Float64Index([900.0, 740.0, 620.0, 500.0, 375.0, 245.0, 90.0], dtype='float64', name='cosp_prs'))
- cosp_tauPandasIndex
PandasIndex(Float64Index([0.15, 0.8, 2.45, 6.5, 16.2, 41.5, 219.5], dtype='float64', name='cosp_tau'))
- cosp_scolPandasIndex
PandasIndex(Int64Index([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50], dtype='int64', name='cosp_scol')) - cosp_htPandasIndex
PandasIndex(Float64Index([ 240.0, 720.0, 1200.0, 1680.0, 2160.0, 2640.0, 3120.0, 3600.0, 4080.0, 4560.0, 5040.0, 5520.0, 6000.0, 6480.0, 6960.0, 7440.0, 7920.0, 8400.0, 8880.0, 9360.0, 9840.0, 10320.0, 10800.0, 11280.0, 11760.0, 12240.0, 12720.0, 13200.0, 13680.0, 14160.0, 14640.0, 15120.0, 15600.0, 16080.0, 16560.0, 17040.0, 17520.0, 18000.0, 18480.0, 18960.0], dtype='float64', name='cosp_ht')) - cosp_srPandasIndex
PandasIndex(Float64Index([ 0.605, 2.1, 4.0, 6.0, 8.5, 12.5, 17.5, 22.5, 27.5, 35.0, 45.0, 55.0, 70.0, 539.5, 1004.0], dtype='float64', name='cosp_sr')) - cosp_szaPandasIndex
PandasIndex(Float64Index([0.0, 15.0, 30.0, 45.0, 60.0], dtype='float64', name='cosp_sza'))
- timePandasIndex
PandasIndex(CFTimeIndex([1920-01-01 00:00:00, 1920-01-01 06:00:00, 1920-01-01 12:00:00, 1920-01-01 18:00:00, 1920-01-02 00:00:00, 1920-01-02 06:00:00, 1920-01-02 12:00:00, 1920-01-02 18:00:00, 1920-01-03 00:00:00, 1920-01-03 06:00:00, ... 1929-12-29 12:00:00, 1929-12-29 18:00:00, 1929-12-30 00:00:00, 1929-12-30 06:00:00, 1929-12-30 12:00:00, 1929-12-30 18:00:00, 1929-12-31 00:00:00, 1929-12-31 06:00:00, 1929-12-31 12:00:00, 1929-12-31 18:00:00], dtype='object', length=14600, calendar='noleap', freq='6H'))
- np :
- 4
- ne :
- 120
- Conventions :
- CF-1.0
- source :
- CAM
- case :
- b.e13.BHISTC5.ne120_t12.cesm-ihesp-hires1.0.30-1920-2100.002
- title :
- UNSET
- logname :
- nanr
- host :
- login1.frontera.
- Version :
- $Name$
- revision_Id :
- $Id$
- initial_file :
- B.E.13.BHISTC5.ne120_t12.sehires38.003.sunway.cam.i.1920-01-01-00000.nc
- topography_file :
- /work/02503/edwardsj/CESM/inputdata//atm/cam/topo/USGS-gtopo30_ne120np4_16xdel2-PFC-consistentSGH.nc
Now to apply the regridder on updated we rename dummy to lat (both are size-1 in updated and dummy_in), and ncol to lon (both are the same size in updated and dummy_in)
regridded = regridder(updated.rename({"dummy": "lat", "ncol": "lon"}))
regridded
<xarray.Dataset>
Dimensions: (lat: 768, lon: 1152, time: 14600, lev: 30, ilev: 31,
cosp_prs: 7, cosp_tau: 7, cosp_scol: 50, cosp_ht: 40,
cosp_sr: 15, cosp_sza: 5)
Coordinates:
* lat (lat) float64 -90.0 -89.77 -89.53 -89.3 ... 89.3 89.53 89.77 90.0
* lon (lon) float64 0.0 0.3125 0.625 0.9375 ... 358.8 359.1 359.4 359.7
* lev (lev) float64 3.643 7.595 14.36 24.61 ... 936.2 957.5 976.3 992.6
* ilev (ilev) float64 2.255 5.032 10.16 18.56 ... 967.5 985.1 1e+03
* cosp_prs (cosp_prs) float64 900.0 740.0 620.0 500.0 375.0 245.0 90.0
* cosp_tau (cosp_tau) float64 0.15 0.8 2.45 6.5 16.2 41.5 219.5
* cosp_scol (cosp_scol) int32 1 2 3 4 5 6 7 8 9 ... 43 44 45 46 47 48 49 50
* cosp_ht (cosp_ht) float64 240.0 720.0 1.2e+03 ... 1.848e+04 1.896e+04
* cosp_sr (cosp_sr) float64 0.605 2.1 4.0 6.0 ... 55.0 70.0 539.5 1.004e+03
* cosp_sza (cosp_sza) float64 0.0 15.0 30.0 45.0 60.0
* time (time) object 1920-01-01 00:00:00 ... 1929-12-31 18:00:00
Data variables:
area (lat, lon) float64 dask.array<chunksize=(768, 1152), meta=np.ndarray>
IVT (time, lat, lon) float32 dask.array<chunksize=(50, 768, 1152), meta=np.ndarray>
Attributes:
regrid_method: bilinear- lat: 768
- lon: 1152
- time: 14600
- lev: 30
- ilev: 31
- cosp_prs: 7
- cosp_tau: 7
- cosp_scol: 50
- cosp_ht: 40
- cosp_sr: 15
- cosp_sza: 5
- lat(lat)float64-90.0 -89.77 -89.53 ... 89.77 90.0
array([-90. , -89.76532, -89.53064, ..., 89.53064, 89.76532, 90. ])
- lon(lon)float640.0 0.3125 0.625 ... 359.4 359.7
array([0.000000e+00, 3.125000e-01, 6.250000e-01, ..., 3.590625e+02, 3.593750e+02, 3.596875e+02]) - lev(lev)float643.643 7.595 14.36 ... 976.3 992.6
array([ 3.643466, 7.59482 , 14.356632, 24.61222 , 38.2683 , 54.59548 , 72.012451, 87.82123 , 103.317127, 121.547241, 142.994039, 168.22508 , 197.908087, 232.828619, 273.910817, 322.241902, 379.100904, 445.992574, 524.687175, 609.778695, 691.38943 , 763.404481, 820.858369, 859.534767, 887.020249, 912.644547, 936.198398, 957.48548 , 976.325407, 992.556095]) - ilev(ilev)float642.255 5.032 10.16 ... 985.1 1e+03
array([ 2.25524 , 5.031692, 10.157947, 18.555317, 30.669123, 45.867477, 63.323483, 80.701418, 94.941042, 111.693211, 131.401271, 154.586807, 181.863353, 213.952821, 251.704417, 296.117216, 348.366588, 409.835219, 482.149929, 567.224421, 652.332969, 730.445892, 796.363071, 845.353667, 873.715866, 900.324631, 924.964462, 947.432335, 967.538625, 985.11219 , 1000. ]) - cosp_prs(cosp_prs)float64900.0 740.0 620.0 ... 245.0 90.0
array([900., 740., 620., 500., 375., 245., 90.])
- cosp_tau(cosp_tau)float640.15 0.8 2.45 6.5 16.2 41.5 219.5
array([1.500e-01, 8.000e-01, 2.450e+00, 6.500e+00, 1.620e+01, 4.150e+01, 2.195e+02]) - cosp_scol(cosp_scol)int321 2 3 4 5 6 7 ... 45 46 47 48 49 50
array([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50], dtype=int32) - cosp_ht(cosp_ht)float64240.0 720.0 ... 1.848e+04 1.896e+04
array([ 240., 720., 1200., 1680., 2160., 2640., 3120., 3600., 4080., 4560., 5040., 5520., 6000., 6480., 6960., 7440., 7920., 8400., 8880., 9360., 9840., 10320., 10800., 11280., 11760., 12240., 12720., 13200., 13680., 14160., 14640., 15120., 15600., 16080., 16560., 17040., 17520., 18000., 18480., 18960.]) - cosp_sr(cosp_sr)float640.605 2.1 4.0 ... 539.5 1.004e+03
array([6.050e-01, 2.100e+00, 4.000e+00, 6.000e+00, 8.500e+00, 1.250e+01, 1.750e+01, 2.250e+01, 2.750e+01, 3.500e+01, 4.500e+01, 5.500e+01, 7.000e+01, 5.395e+02, 1.004e+03]) - cosp_sza(cosp_sza)float640.0 15.0 30.0 45.0 60.0
array([ 0., 15., 30., 45., 60.])
- time(time)object1920-01-01 00:00:00 ... 1929-12-...
array([cftime.DatetimeNoLeap(1920, 1, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1920, 1, 1, 6, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1920, 1, 1, 12, 0, 0, 0, has_year_zero=True), ..., cftime.DatetimeNoLeap(1929, 12, 31, 6, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1929, 12, 31, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1929, 12, 31, 18, 0, 0, 0, has_year_zero=True)], dtype=object)
- area(lat, lon)float64dask.array<chunksize=(768, 1152), meta=np.ndarray>
Array Chunk Bytes 6.75 MiB 6.75 MiB Shape (768, 1152) (768, 1152) Dask graph 1 chunks in 6 graph layers Data type float64 numpy.ndarray - IVT(time, lat, lon)float32dask.array<chunksize=(50, 768, 1152), meta=np.ndarray>
Array Chunk Bytes 48.12 GiB 168.75 MiB Shape (14600, 768, 1152) (50, 768, 1152) Dask graph 292 chunks in 7 graph layers Data type float32 numpy.ndarray
- levPandasIndex
PandasIndex(Float64Index([ 3.64346569404006, 7.594819646328688, 14.356632251292467, 24.612220004200935, 38.26829977333546, 54.59547974169254, 72.01245054602623, 87.82123029232025, 103.31712663173676, 121.54724076390266, 142.99403876066208, 168.22507977485657, 197.9080867022276, 232.82861895859241, 273.9108167588711, 322.2419023513794, 379.10090386867523, 445.992574095726, 524.6871747076511, 609.7786948084831, 691.3894303143024, 763.404481112957, 820.8583686500788, 859.5347665250301, 887.0202489197254, 912.644546944648, 936.1983984708786, 957.485479535535, 976.325407391414, 992.556095123291], dtype='float64', name='lev')) - ilevPandasIndex
PandasIndex(Float64Index([ 2.255239523947239, 5.031691864132881, 10.15794742852449, 18.55531707406044, 30.66912293434143, 45.86747661232948, 63.3234828710556, 80.70141822099686, 94.94104236364365, 111.69321089982991, 131.4012706279755, 154.5868068933487, 181.8633526563644, 213.95282074809072, 251.70441716909409, 296.11721634864807, 348.3665883541107, 409.8352193832398, 482.14992880821234, 567.22442060709, 652.3329690098763, 730.4458916187286, 796.3630706071854, 845.3536666929722, 873.7158663570881, 900.3246314823627, 924.9644624069333, 947.4323345348239, 967.5386245362461, 985.112190246582, 1000.0], dtype='float64', name='ilev')) - cosp_prsPandasIndex
PandasIndex(Float64Index([900.0, 740.0, 620.0, 500.0, 375.0, 245.0, 90.0], dtype='float64', name='cosp_prs'))
- cosp_tauPandasIndex
PandasIndex(Float64Index([0.15, 0.8, 2.45, 6.5, 16.2, 41.5, 219.5], dtype='float64', name='cosp_tau'))
- cosp_scolPandasIndex
PandasIndex(Int64Index([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50], dtype='int64', name='cosp_scol')) - cosp_htPandasIndex
PandasIndex(Float64Index([ 240.0, 720.0, 1200.0, 1680.0, 2160.0, 2640.0, 3120.0, 3600.0, 4080.0, 4560.0, 5040.0, 5520.0, 6000.0, 6480.0, 6960.0, 7440.0, 7920.0, 8400.0, 8880.0, 9360.0, 9840.0, 10320.0, 10800.0, 11280.0, 11760.0, 12240.0, 12720.0, 13200.0, 13680.0, 14160.0, 14640.0, 15120.0, 15600.0, 16080.0, 16560.0, 17040.0, 17520.0, 18000.0, 18480.0, 18960.0], dtype='float64', name='cosp_ht')) - cosp_srPandasIndex
PandasIndex(Float64Index([ 0.605, 2.1, 4.0, 6.0, 8.5, 12.5, 17.5, 22.5, 27.5, 35.0, 45.0, 55.0, 70.0, 539.5, 1004.0], dtype='float64', name='cosp_sr')) - cosp_szaPandasIndex
PandasIndex(Float64Index([0.0, 15.0, 30.0, 45.0, 60.0], dtype='float64', name='cosp_sza'))
- timePandasIndex
PandasIndex(CFTimeIndex([1920-01-01 00:00:00, 1920-01-01 06:00:00, 1920-01-01 12:00:00, 1920-01-01 18:00:00, 1920-01-02 00:00:00, 1920-01-02 06:00:00, 1920-01-02 12:00:00, 1920-01-02 18:00:00, 1920-01-03 00:00:00, 1920-01-03 06:00:00, ... 1929-12-29 12:00:00, 1929-12-29 18:00:00, 1929-12-30 00:00:00, 1929-12-30 06:00:00, 1929-12-30 12:00:00, 1929-12-30 18:00:00, 1929-12-31 00:00:00, 1929-12-31 06:00:00, 1929-12-31 12:00:00, 1929-12-31 18:00:00], dtype='object', length=14600, calendar='noleap', freq='6H')) - latPandasIndex
PandasIndex(Float64Index([ -90.0, -89.76531982421875, -89.5306396484375, -89.29595947265625, -89.061279296875, -88.82659912109375, -88.5919189453125, -88.35723876953125, -88.12255859375, -87.88787841796875, ... 87.88787841796875, 88.12255859375, 88.35723876953125, 88.5919189453125, 88.82659912109375, 89.061279296875, 89.29595947265625, 89.5306396484375, 89.76531982421875, 90.0], dtype='float64', name='lat', length=768)) - lonPandasIndex
PandasIndex(Float64Index([ 0.0, 0.3125, 0.625, 0.9375, 1.25, 1.5625, 1.875, 2.1875, 2.5, 2.8125, ... 356.875, 357.1875, 357.5, 357.8125, 358.125, 358.4375, 358.75, 359.0625, 359.375, 359.6875], dtype='float64', name='lon', length=1152))
- regrid_method :
- bilinear
Visualize#
Here we’ll visualize a single timestep but note that regridded.IVT.mean("time") will nicely parallelize with dask.
figure = regridded.IVT.isel(time=100).hvplot(
cmap="twilight", clim=(0, 1000), frame_width=500, geo=True, coastline=True
)
figure
Wrap it up#
def regrid_cam_se(dataset, weight_file):
"""
Regrid CAM-SE output using an existing ESMF weights file.
Parameters
----------
dataset: xarray.Dataset
Input dataset to be regridded. Must have the `ncol` dimension.
weight_file: str or Path
Path to existing ESMF weights file
Returns
-------
regridded
xarray.Dataset after regridding.
"""
import numpy as np
import xarray as xr
assert isinstance(dataset, xr.Dataset)
weights = xr.open_dataset(weight_file)
# input variable shape
in_shape = weights.src_grid_dims.load().data
# Since xESMF expects 2D vars, we'll insert a dummy dimension of size-1
if len(in_shape) == 1:
in_shape = [1, in_shape.item()]
# output variable shape
out_shape = weights.dst_grid_dims.load().data.tolist()[::-1]
print(f"Regridding from {in_shape} to {out_shape}")
# Insert dummy dimension
vars_with_ncol = [name for name in data_in.variables if "ncol" in data_in[name].dims]
updated = data_in.copy().update(
data_in[vars_with_ncol].transpose(..., "ncol").expand_dims("dummy", axis=-2)
)
# Actually regrid, after renaming
regridded = regridder(updated.rename({"dummy": "lat", "ncol": "lon"}))
# merge back any variables that didn't have the ncol dimension
# And so were not regridded
return xr.merge([data_in.drop_vars(regridded.variables), regridded])
regrid_cam_se(data_in, weight_file)
Regridding from [1, 777602] to [768, 1152]
<xarray.Dataset>
Dimensions: (lev: 30, ilev: 31, cosp_prs: 7, nbnd: 2, cosp_tau: 7,
cosp_ht: 40, cosp_sr: 15, time: 14600, lat: 768, lon: 1152,
cosp_scol: 50, cosp_sza: 5)
Coordinates:
* lev (lev) float64 3.643 7.595 14.36 24.61 ... 957.5 976.3 992.6
* ilev (ilev) float64 2.255 5.032 10.16 18.56 ... 967.5 985.1 1e+03
* cosp_prs (cosp_prs) float64 900.0 740.0 620.0 500.0 375.0 245.0 90.0
* cosp_tau (cosp_tau) float64 0.15 0.8 2.45 6.5 16.2 41.5 219.5
* cosp_ht (cosp_ht) float64 240.0 720.0 1.2e+03 ... 1.848e+04 1.896e+04
* cosp_sr (cosp_sr) float64 0.605 2.1 4.0 6.0 ... 70.0 539.5 1.004e+03
* time (time) object 1920-01-01 00:00:00 ... 1929-12-31 18:00:00
* lat (lat) float64 -90.0 -89.77 -89.53 -89.3 ... 89.53 89.77 90.0
* lon (lon) float64 0.0 0.3125 0.625 0.9375 ... 359.1 359.4 359.7
* cosp_scol (cosp_scol) int32 1 2 3 4 5 6 7 8 ... 43 44 45 46 47 48 49 50
* cosp_sza (cosp_sza) float64 0.0 15.0 30.0 45.0 60.0
Dimensions without coordinates: nbnd
Data variables: (12/33)
hyam (lev) float64 dask.array<chunksize=(30,), meta=np.ndarray>
hybm (lev) float64 dask.array<chunksize=(30,), meta=np.ndarray>
P0 float64 ...
hyai (ilev) float64 dask.array<chunksize=(31,), meta=np.ndarray>
hybi (ilev) float64 dask.array<chunksize=(31,), meta=np.ndarray>
cosp_prs_bnds (cosp_prs, nbnd) float64 dask.array<chunksize=(7, 2), meta=np.ndarray>
... ...
f11vmr (time) float64 dask.array<chunksize=(50,), meta=np.ndarray>
f12vmr (time) float64 dask.array<chunksize=(50,), meta=np.ndarray>
sol_tsi (time) float64 dask.array<chunksize=(50,), meta=np.ndarray>
nsteph (time) int32 dask.array<chunksize=(50,), meta=np.ndarray>
area (lat, lon) float64 dask.array<chunksize=(768, 1152), meta=np.ndarray>
IVT (time, lat, lon) float32 dask.array<chunksize=(50, 768, 1152), meta=np.ndarray>
Attributes:
np: 4
ne: 120
Conventions: CF-1.0
source: CAM
case: b.e13.BHISTC5.ne120_t12.cesm-ihesp-hires1.0.30-1920-210...
title: UNSET
logname: nanr
host: login1.frontera.
Version: $Name$
revision_Id: $Id$
initial_file: B.E.13.BHISTC5.ne120_t12.sehires38.003.sunway.cam.i.192...
topography_file: /work/02503/edwardsj/CESM/inputdata//atm/cam/topo/USGS-...- lev: 30
- ilev: 31
- cosp_prs: 7
- nbnd: 2
- cosp_tau: 7
- cosp_ht: 40
- cosp_sr: 15
- time: 14600
- lat: 768
- lon: 1152
- cosp_scol: 50
- cosp_sza: 5
- lev(lev)float643.643 7.595 14.36 ... 976.3 992.6
array([ 3.643466, 7.59482 , 14.356632, 24.61222 , 38.2683 , 54.59548 , 72.012451, 87.82123 , 103.317127, 121.547241, 142.994039, 168.22508 , 197.908087, 232.828619, 273.910817, 322.241902, 379.100904, 445.992574, 524.687175, 609.778695, 691.38943 , 763.404481, 820.858369, 859.534767, 887.020249, 912.644547, 936.198398, 957.48548 , 976.325407, 992.556095]) - ilev(ilev)float642.255 5.032 10.16 ... 985.1 1e+03
array([ 2.25524 , 5.031692, 10.157947, 18.555317, 30.669123, 45.867477, 63.323483, 80.701418, 94.941042, 111.693211, 131.401271, 154.586807, 181.863353, 213.952821, 251.704417, 296.117216, 348.366588, 409.835219, 482.149929, 567.224421, 652.332969, 730.445892, 796.363071, 845.353667, 873.715866, 900.324631, 924.964462, 947.432335, 967.538625, 985.11219 , 1000. ]) - cosp_prs(cosp_prs)float64900.0 740.0 620.0 ... 245.0 90.0
array([900., 740., 620., 500., 375., 245., 90.])
- cosp_tau(cosp_tau)float640.15 0.8 2.45 6.5 16.2 41.5 219.5
array([1.500e-01, 8.000e-01, 2.450e+00, 6.500e+00, 1.620e+01, 4.150e+01, 2.195e+02]) - cosp_ht(cosp_ht)float64240.0 720.0 ... 1.848e+04 1.896e+04
array([ 240., 720., 1200., 1680., 2160., 2640., 3120., 3600., 4080., 4560., 5040., 5520., 6000., 6480., 6960., 7440., 7920., 8400., 8880., 9360., 9840., 10320., 10800., 11280., 11760., 12240., 12720., 13200., 13680., 14160., 14640., 15120., 15600., 16080., 16560., 17040., 17520., 18000., 18480., 18960.]) - cosp_sr(cosp_sr)float640.605 2.1 4.0 ... 539.5 1.004e+03
array([6.050e-01, 2.100e+00, 4.000e+00, 6.000e+00, 8.500e+00, 1.250e+01, 1.750e+01, 2.250e+01, 2.750e+01, 3.500e+01, 4.500e+01, 5.500e+01, 7.000e+01, 5.395e+02, 1.004e+03]) - time(time)object1920-01-01 00:00:00 ... 1929-12-...
array([cftime.DatetimeNoLeap(1920, 1, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1920, 1, 1, 6, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1920, 1, 1, 12, 0, 0, 0, has_year_zero=True), ..., cftime.DatetimeNoLeap(1929, 12, 31, 6, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1929, 12, 31, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1929, 12, 31, 18, 0, 0, 0, has_year_zero=True)], dtype=object) - lat(lat)float64-90.0 -89.77 -89.53 ... 89.77 90.0
array([-90. , -89.76532, -89.53064, ..., 89.53064, 89.76532, 90. ])
- lon(lon)float640.0 0.3125 0.625 ... 359.4 359.7
array([0.000000e+00, 3.125000e-01, 6.250000e-01, ..., 3.590625e+02, 3.593750e+02, 3.596875e+02]) - cosp_scol(cosp_scol)int321 2 3 4 5 6 7 ... 45 46 47 48 49 50
array([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50], dtype=int32) - cosp_sza(cosp_sza)float640.0 15.0 30.0 45.0 60.0
array([ 0., 15., 30., 45., 60.])
- hyam(lev)float64dask.array<chunksize=(30,), meta=np.ndarray>
- long_name :
- hybrid A coefficient at layer midpoints
Array Chunk Bytes 240 B 240 B Shape (30,) (30,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - hybm(lev)float64dask.array<chunksize=(30,), meta=np.ndarray>
- long_name :
- hybrid B coefficient at layer midpoints
Array Chunk Bytes 240 B 240 B Shape (30,) (30,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - P0()float64...
- long_name :
- reference pressure
- units :
- Pa
[1 values with dtype=float64]
- hyai(ilev)float64dask.array<chunksize=(31,), meta=np.ndarray>
- long_name :
- hybrid A coefficient at layer interfaces
Array Chunk Bytes 248 B 248 B Shape (31,) (31,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - hybi(ilev)float64dask.array<chunksize=(31,), meta=np.ndarray>
- long_name :
- hybrid B coefficient at layer interfaces
Array Chunk Bytes 248 B 248 B Shape (31,) (31,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - cosp_prs_bnds(cosp_prs, nbnd)float64dask.array<chunksize=(7, 2), meta=np.ndarray>
Array Chunk Bytes 112 B 112 B Shape (7, 2) (7, 2) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - cosp_tau_bnds(cosp_tau, nbnd)float64dask.array<chunksize=(7, 2), meta=np.ndarray>
Array Chunk Bytes 112 B 112 B Shape (7, 2) (7, 2) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - cosp_ht_bnds(cosp_ht, nbnd)float64dask.array<chunksize=(40, 2), meta=np.ndarray>
Array Chunk Bytes 640 B 640 B Shape (40, 2) (40, 2) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - cosp_sr_bnds(cosp_sr, nbnd)float64dask.array<chunksize=(15, 2), meta=np.ndarray>
Array Chunk Bytes 240 B 240 B Shape (15, 2) (15, 2) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - ntrm()int32...
- long_name :
- spectral truncation parameter M
[1 values with dtype=int32]
- ntrn()int32...
- long_name :
- spectral truncation parameter N
[1 values with dtype=int32]
- ntrk()int32...
- long_name :
- spectral truncation parameter K
[1 values with dtype=int32]
- ndbase()int32...
- long_name :
- base day
[1 values with dtype=int32]
- nsbase()int32...
- long_name :
- seconds of base day
[1 values with dtype=int32]
- nbdate()int32...
- long_name :
- base date (YYYYMMDD)
[1 values with dtype=int32]
- nbsec()int32...
- long_name :
- seconds of base date
[1 values with dtype=int32]
- mdt()int32...
- long_name :
- timestep
- units :
- s
[1 values with dtype=int32]
- date(time)int32dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- current date (YYYYMMDD)
Array Chunk Bytes 57.03 kiB 200 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type int32 numpy.ndarray - datesec(time)int32dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- current seconds of current date
Array Chunk Bytes 57.03 kiB 200 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type int32 numpy.ndarray - time_bnds(time, nbnd)objectdask.array<chunksize=(50, 2), meta=np.ndarray>
- long_name :
- time interval endpoints
Array Chunk Bytes 228.12 kiB 800 B Shape (14600, 2) (50, 2) Dask graph 292 chunks in 2 graph layers Data type object numpy.ndarray - date_written(time)|S8dask.array<chunksize=(50,), meta=np.ndarray>
Array Chunk Bytes 114.06 kiB 400 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type |S8 numpy.ndarray - time_written(time)|S8dask.array<chunksize=(50,), meta=np.ndarray>
Array Chunk Bytes 114.06 kiB 400 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type |S8 numpy.ndarray - ndcur(time)int32dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- current day (from base day)
Array Chunk Bytes 57.03 kiB 200 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type int32 numpy.ndarray - nscur(time)int32dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- current seconds of current day
Array Chunk Bytes 57.03 kiB 200 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type int32 numpy.ndarray - co2vmr(time)float64dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- co2 volume mixing ratio
Array Chunk Bytes 114.06 kiB 400 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type float64 numpy.ndarray - ch4vmr(time)float64dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- ch4 volume mixing ratio
Array Chunk Bytes 114.06 kiB 400 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type float64 numpy.ndarray - n2ovmr(time)float64dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- n2o volume mixing ratio
Array Chunk Bytes 114.06 kiB 400 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type float64 numpy.ndarray - f11vmr(time)float64dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- f11 volume mixing ratio
Array Chunk Bytes 114.06 kiB 400 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type float64 numpy.ndarray - f12vmr(time)float64dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- f12 volume mixing ratio
Array Chunk Bytes 114.06 kiB 400 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type float64 numpy.ndarray - sol_tsi(time)float64dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- total solar irradiance
- units :
- W/m2
Array Chunk Bytes 114.06 kiB 400 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type float64 numpy.ndarray - nsteph(time)int32dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- current timestep
Array Chunk Bytes 57.03 kiB 200 B Shape (14600,) (50,) Dask graph 292 chunks in 2 graph layers Data type int32 numpy.ndarray - area(lat, lon)float64dask.array<chunksize=(768, 1152), meta=np.ndarray>
Array Chunk Bytes 6.75 MiB 6.75 MiB Shape (768, 1152) (768, 1152) Dask graph 1 chunks in 6 graph layers Data type float64 numpy.ndarray - IVT(time, lat, lon)float32dask.array<chunksize=(50, 768, 1152), meta=np.ndarray>
Array Chunk Bytes 48.12 GiB 168.75 MiB Shape (14600, 768, 1152) (50, 768, 1152) Dask graph 292 chunks in 7 graph layers Data type float32 numpy.ndarray
- levPandasIndex
PandasIndex(Float64Index([ 3.64346569404006, 7.594819646328688, 14.356632251292467, 24.612220004200935, 38.26829977333546, 54.59547974169254, 72.01245054602623, 87.82123029232025, 103.31712663173676, 121.54724076390266, 142.99403876066208, 168.22507977485657, 197.9080867022276, 232.82861895859241, 273.9108167588711, 322.2419023513794, 379.10090386867523, 445.992574095726, 524.6871747076511, 609.7786948084831, 691.3894303143024, 763.404481112957, 820.8583686500788, 859.5347665250301, 887.0202489197254, 912.644546944648, 936.1983984708786, 957.485479535535, 976.325407391414, 992.556095123291], dtype='float64', name='lev')) - ilevPandasIndex
PandasIndex(Float64Index([ 2.255239523947239, 5.031691864132881, 10.15794742852449, 18.55531707406044, 30.66912293434143, 45.86747661232948, 63.3234828710556, 80.70141822099686, 94.94104236364365, 111.69321089982991, 131.4012706279755, 154.5868068933487, 181.8633526563644, 213.95282074809072, 251.70441716909409, 296.11721634864807, 348.3665883541107, 409.8352193832398, 482.14992880821234, 567.22442060709, 652.3329690098763, 730.4458916187286, 796.3630706071854, 845.3536666929722, 873.7158663570881, 900.3246314823627, 924.9644624069333, 947.4323345348239, 967.5386245362461, 985.112190246582, 1000.0], dtype='float64', name='ilev')) - cosp_prsPandasIndex
PandasIndex(Float64Index([900.0, 740.0, 620.0, 500.0, 375.0, 245.0, 90.0], dtype='float64', name='cosp_prs'))
- cosp_tauPandasIndex
PandasIndex(Float64Index([0.15, 0.8, 2.45, 6.5, 16.2, 41.5, 219.5], dtype='float64', name='cosp_tau'))
- cosp_htPandasIndex
PandasIndex(Float64Index([ 240.0, 720.0, 1200.0, 1680.0, 2160.0, 2640.0, 3120.0, 3600.0, 4080.0, 4560.0, 5040.0, 5520.0, 6000.0, 6480.0, 6960.0, 7440.0, 7920.0, 8400.0, 8880.0, 9360.0, 9840.0, 10320.0, 10800.0, 11280.0, 11760.0, 12240.0, 12720.0, 13200.0, 13680.0, 14160.0, 14640.0, 15120.0, 15600.0, 16080.0, 16560.0, 17040.0, 17520.0, 18000.0, 18480.0, 18960.0], dtype='float64', name='cosp_ht')) - cosp_srPandasIndex
PandasIndex(Float64Index([ 0.605, 2.1, 4.0, 6.0, 8.5, 12.5, 17.5, 22.5, 27.5, 35.0, 45.0, 55.0, 70.0, 539.5, 1004.0], dtype='float64', name='cosp_sr')) - timePandasIndex
PandasIndex(CFTimeIndex([1920-01-01 00:00:00, 1920-01-01 06:00:00, 1920-01-01 12:00:00, 1920-01-01 18:00:00, 1920-01-02 00:00:00, 1920-01-02 06:00:00, 1920-01-02 12:00:00, 1920-01-02 18:00:00, 1920-01-03 00:00:00, 1920-01-03 06:00:00, ... 1929-12-29 12:00:00, 1929-12-29 18:00:00, 1929-12-30 00:00:00, 1929-12-30 06:00:00, 1929-12-30 12:00:00, 1929-12-30 18:00:00, 1929-12-31 00:00:00, 1929-12-31 06:00:00, 1929-12-31 12:00:00, 1929-12-31 18:00:00], dtype='object', length=14600, calendar='noleap', freq='6H')) - latPandasIndex
PandasIndex(Float64Index([ -90.0, -89.76531982421875, -89.5306396484375, -89.29595947265625, -89.061279296875, -88.82659912109375, -88.5919189453125, -88.35723876953125, -88.12255859375, -87.88787841796875, ... 87.88787841796875, 88.12255859375, 88.35723876953125, 88.5919189453125, 88.82659912109375, 89.061279296875, 89.29595947265625, 89.5306396484375, 89.76531982421875, 90.0], dtype='float64', name='lat', length=768)) - lonPandasIndex
PandasIndex(Float64Index([ 0.0, 0.3125, 0.625, 0.9375, 1.25, 1.5625, 1.875, 2.1875, 2.5, 2.8125, ... 356.875, 357.1875, 357.5, 357.8125, 358.125, 358.4375, 358.75, 359.0625, 359.375, 359.6875], dtype='float64', name='lon', length=1152)) - cosp_scolPandasIndex
PandasIndex(Int64Index([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50], dtype='int64', name='cosp_scol')) - cosp_szaPandasIndex
PandasIndex(Float64Index([0.0, 15.0, 30.0, 45.0, 60.0], dtype='float64', name='cosp_sza'))
- np :
- 4
- ne :
- 120
- Conventions :
- CF-1.0
- source :
- CAM
- case :
- b.e13.BHISTC5.ne120_t12.cesm-ihesp-hires1.0.30-1920-2100.002
- title :
- UNSET
- logname :
- nanr
- host :
- login1.frontera.
- Version :
- $Name$
- revision_Id :
- $Id$
- initial_file :
- B.E.13.BHISTC5.ne120_t12.sehires38.003.sunway.cam.i.1920-01-01-00000.nc
- topography_file :
- /work/02503/edwardsj/CESM/inputdata//atm/cam/topo/USGS-gtopo30_ne120np4_16xdel2-PFC-consistentSGH.nc
Possible improvements#
It should be possible for xESMF to do all this internally and allow the user to create a Regridder as
xesmf.Regridder.from_weights_file(
# path to weights file
path=...,
# input dimension names that will be removed while regridding
# These are "core dimensions"
dims_in=("ncol",),
# Output dataset with lat, lon information
ds_out=...,
)
The repr will need to be updated to show the proper weights path, and input/output core dimensions and variables.