Reading WRF data into Xarray and Visualizing the Output using hvPlot#
The typical data workflow within the Python ecosystem when working with Weather Research and Forecasting (WRF) data is to use the wrf-python package! Traditionally, it can be difficult to utilize the xarray data model with WRF data, due to a few challenges:
WRF data not being CF-compliant (which makes it hard for xarray to properly construct the dataset out of the box using xr.open_dataset)
wrf-python requiring to interact with both netCDF4-python and xarray’s APIs (which can be a daunting task)
The lack of functionality in wrf-python needed to take full advantage of dask’s laziness and parallelism
In this example, we show how you can use the extremely experimental package xWRF, to read in data and plot an interactive visualization of your data!
Again, the stress here is experimental such that this is a proof of concept - not meant to be used directly in workflows; but rather to show what is possible given further development.
By the end of this example, we will generate an interactive plot which looks like the following!
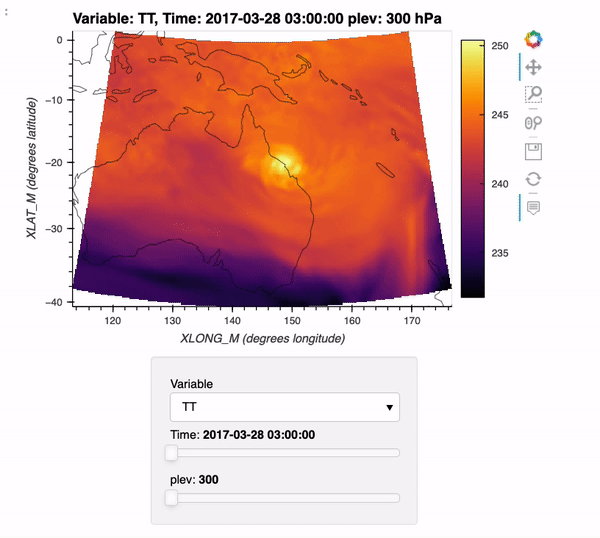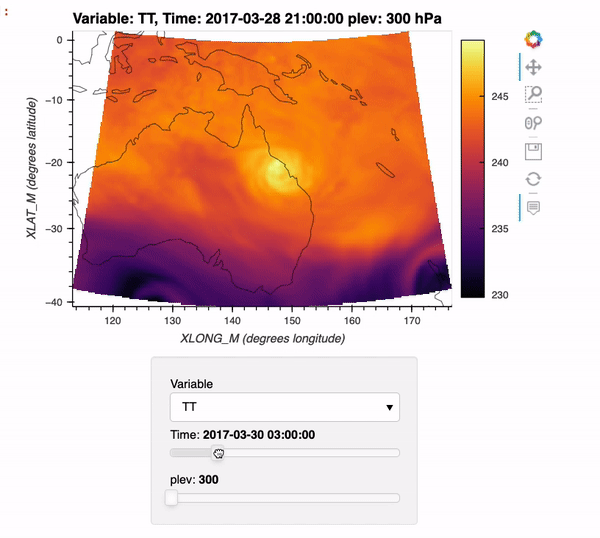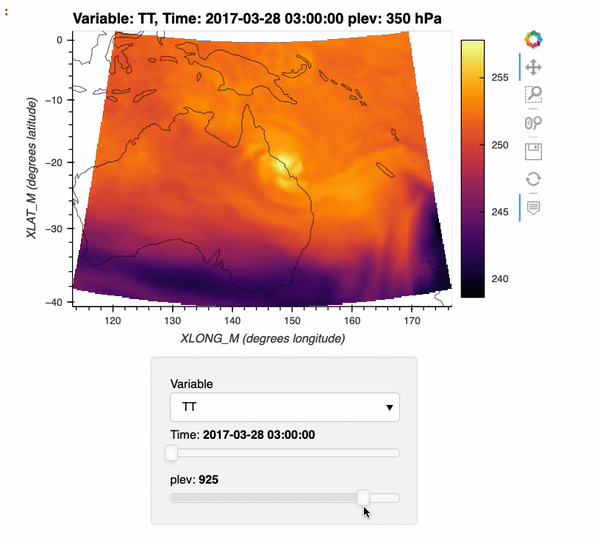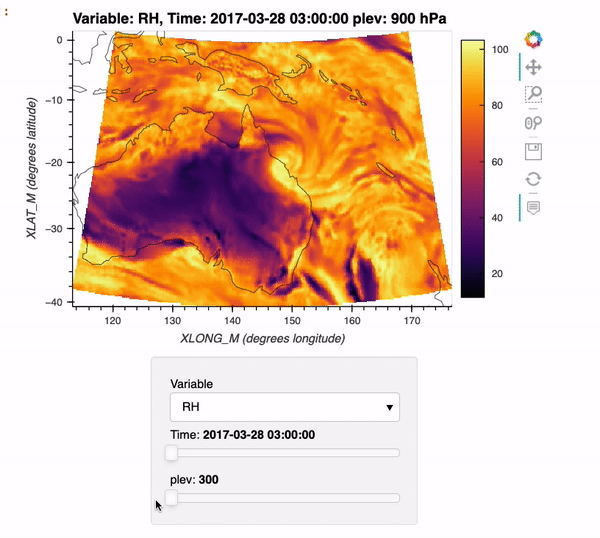
Installing xwrf#
Before we start using xwrf, we need to install it. We can install by following these steps!
Install this in your python environment using pip (
pip install git+https://github.com/NCAR/xwrf.git)Open up a notebook and use the imports shown below!
What exactly is xwrf?#
xwrf provides an xarray backend, which helps with reading in the file. When you are working with non-cf-compliant datasets (ex. WRF output), this backends transform the file into a format that is easier to work with (ex. helpful coordinate information).
You could complete this task using a preprocess function (see xarray open_mfdataset documentation), but this would get tricky once you add more advanced IO functionality.
Imports#
Here, we only need a few packages; xwrf, dask, hvplot/holoviews, and xarray
import glob
import holoviews as hv
import hvplot
import hvplot.xarray
import xarray as xr
import xwrf
from distributed import Client
from ncar_jobqueue import NCARCluster
hv.extension('bokeh')
Spin up a Cluster#
cluster = NCARCluster()
cluster.scale(10)
client = Client(cluster)
client
Client
Client-84daf380-2df5-11ec-976e-3cecef1acbfa
| Connection method: Cluster object | Cluster type: PBSCluster |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/mgrover/proxy/8787/status |
Cluster Info
PBSCluster
327558cf
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/mgrover/proxy/8787/status | Workers: 0 |
| Total threads: 0 | Total memory: 0 B |
Scheduler Info
Scheduler
Scheduler-d1280585-a386-4383-acb9-63ea4f6b110e
| Comm: tcp://10.12.206.63:34961 | Workers: 0 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/mgrover/proxy/8787/status | Total threads: 0 |
| Started: Just now | Total memory: 0 B |
Workers
Grab a list of files#
We are using some sample data provided by Cindy Bruyere, who works in the Mesoscale & Microscale Meteorology Lab at NCAR! She proposed this challenge of finding a more performant way of getting WRF data into the Xarray data model. The data here is from a regional model simulation over Australia!
files = sorted(glob.glob('/glade/scratch/bruyerec/IAG/METGRID/*.nc'))
The data are in 3 hourly chunks, with 8 timesteps per day. Let’s grab the last 80 timesteps from the simulation!
file_subset = files[-80:]
Examine of the files#
We can open up one of the files, and inspect which variables are included, as well as descriptions of those variables!
wrf_ds = xr.open_dataset(files[0])
wrf_ds
<xarray.Dataset>
Dimensions: (Time: 1, num_metgrid_levels: 38, south_north: 378, west_east: 480, num_st_layers: 4, num_sm_layers: 4, south_north_stag: 379, west_east_stag: 481, z-dimension0003: 3, z-dimension0132: 132, z-dimension0012: 12, z-dimension0016: 16, z-dimension0021: 21)
Dimensions without coordinates: Time, num_metgrid_levels, south_north, west_east, num_st_layers, num_sm_layers, south_north_stag, west_east_stag, z-dimension0003, z-dimension0132, z-dimension0012, z-dimension0016, z-dimension0021
Data variables: (12/107)
Times (Time) |S19 b'1954-02-17_00:00:00'
PRES (Time, num_metgrid_levels, south_north, west_east) float32 ...
SOIL_LAYERS (Time, num_st_layers, south_north, west_east) float32 ...
SM (Time, num_sm_layers, south_north, west_east) float32 ...
ST (Time, num_st_layers, south_north, west_east) float32 ...
GHT (Time, num_metgrid_levels, south_north, west_east) float32 ...
... ...
XLONG_U (Time, south_north, west_east_stag) float32 ...
XLAT_U (Time, south_north, west_east_stag) float32 ...
XLONG_V (Time, south_north_stag, west_east) float32 ...
XLAT_V (Time, south_north_stag, west_east) float32 ...
XLONG_M (Time, south_north, west_east) float32 ...
XLAT_M (Time, south_north, west_east) float32 ...
Attributes: (12/73)
TITLE: OUTPUT FROM METGRID V4.3
SIMULATION_START_DATE: 1954-02-17_00:00:00
WEST-EAST_GRID_DIMENSION: 481
SOUTH-NORTH_GRID_DIMENSION: 379
BOTTOM-TOP_GRID_DIMENSION: 38
WEST-EAST_PATCH_START_UNSTAG: 1
... ...
FLAG_FRC_URB2D: 1
FLAG_IMPERV: 1
FLAG_CANFRA: 1
FLAG_EROD: 1
FLAG_CLAYFRAC: 1
FLAG_SANDFRAC: 1- Time: 1
- num_metgrid_levels: 38
- south_north: 378
- west_east: 480
- num_st_layers: 4
- num_sm_layers: 4
- south_north_stag: 379
- west_east_stag: 481
- z-dimension0003: 3
- z-dimension0132: 132
- z-dimension0012: 12
- z-dimension0016: 16
- z-dimension0021: 21
- Times(Time)|S19...
array([b'1954-02-17_00:00:00'], dtype='|S19')
- PRES(Time, num_metgrid_levels, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XYZ
- units :
- description :
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[6894720 values with dtype=float32]
- SOIL_LAYERS(Time, num_st_layers, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XYZ
- units :
- description :
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[725760 values with dtype=float32]
- SM(Time, num_sm_layers, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XYZ
- units :
- description :
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[725760 values with dtype=float32]
- ST(Time, num_st_layers, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XYZ
- units :
- description :
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[725760 values with dtype=float32]
- GHT(Time, num_metgrid_levels, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XYZ
- units :
- m
- description :
- Height
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[6894720 values with dtype=float32]
- SM100289(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- m3 m-3
- description :
- Soil moisture of 100-289 cm ground layer
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- SM028100(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- m3 m-3
- description :
- Soil moisture of 28-100 cm ground layer
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- SM007028(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- m3 m-3
- description :
- Soil moisture of 7-28 cm ground layer
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- SM000007(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- m3 m-3
- description :
- Soil moisture of 0-7 cm ground layer
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- ST100289(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- K
- description :
- T of 100-289 cm ground layer
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- ST028100(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- K
- description :
- T of 28-100 cm ground layer
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- ST007028(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- K
- description :
- T of 7-28 cm ground layer
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- ST000007(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- K
- description :
- T of 0-7 cm ground layer
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- SNOWH(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- m
- description :
- Physical Snow Depth
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- SNOW(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- kg m-2
- description :
- Water Equivalent of Accumulated Snow Depth
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- SST(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- K
- description :
- Sea-Surface Temperature
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- SEAICE(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- fraction
- description :
- Sea-Ice Fraction
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- SKINTEMP(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- K
- description :
- Sea-Surface Temperature
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- PMSL(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- Pa
- description :
- Sea-level Pressure
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- PSFC(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- Pa
- description :
- Surface Pressure
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- RH(Time, num_metgrid_levels, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XYZ
- units :
- %
- description :
- Relative Humidity
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[6894720 values with dtype=float32]
- VV(Time, num_metgrid_levels, south_north_stag, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XYZ
- units :
- m s-1
- description :
- V
- stagger :
- V
- sr_x :
- 1
- sr_y :
- 1
[6912960 values with dtype=float32]
- UU(Time, num_metgrid_levels, south_north, west_east_stag)float32...
- FieldType :
- 104
- MemoryOrder :
- XYZ
- units :
- m s-1
- description :
- U
- stagger :
- U
- sr_x :
- 1
- sr_y :
- 1
[6909084 values with dtype=float32]
- TT(Time, num_metgrid_levels, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XYZ
- units :
- K
- description :
- Temperature
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[6894720 values with dtype=float32]
- SOILHGT(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- m
- description :
- Terrain field of source analysis
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- LANDSEA(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- 0/1 Flag
- description :
- Land/Sea flag
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- OL4SS(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- -
- description :
- orographic effective length for North-Westerly flow (small-scale)
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- OL3SS(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- -
- description :
- orographic effective length for South-Westerly flow (small-scale)
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- OL2SS(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- -
- description :
- orographic effective length for Southerly flow (small-scale)
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- OL1SS(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- -
- description :
- orographic effective length for Westerly flow (small-scale)
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- OA4SS(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- -
- description :
- orographic asymmetry in North-West direction (small-scale)
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- OA3SS(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- -
- description :
- orographic asymmetry in South-West direction (small-scale)
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- OA2SS(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- -
- description :
- orographic asymmetry in South direction (small-scale)
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- OA1SS(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- -
- description :
- orographic asymmetry in West direction (small-scale)
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- VARSS(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- meters
- description :
- standard deviation of small-scale topography
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- CONSS(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- -
- description :
- convexity of small-scale topography
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- OL4LS(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- -
- description :
- orographic effective length (detrended) for North-Westerly flow
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- OL3LS(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- -
- description :
- orographic effective length (detrended) for South-Westerly flow
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- OL2LS(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- -
- description :
- orographic effective length (detrended) for Southerly flow
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- OL1LS(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- -
- description :
- orographic effective length (detrended) for Westerly flow
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- OA4LS(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- -
- description :
- orographic asymmetry (detrended) in North-West direction
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- OA3LS(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- -
- description :
- orographic asymmetry (detrended) in South-West direction
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- OA2LS(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- -
- description :
- orographic asymmetry (detrended) in South
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- OA1LS(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- -
- description :
- orographic asymmetry (detrended) in West direction
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- VARLS(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- meters
- description :
- standard deviation of detrended topography
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- CONLS(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- -
- description :
- convexity of detrended topography
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- IRRIGATION(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- percent
- description :
- irrigated land percentage
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- SANDFRAC(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- fraction
- description :
- Sand Fraction
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- CLAYFRAC(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- fraction
- description :
- Clay Fraction
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- EROD(Time, z-dimension0003, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XYZ
- units :
- fraction
- description :
- EROD
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[544320 values with dtype=float32]
- CANFRA(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- Percent
- description :
- 2011 NLCD Canopy Percent
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- IMPERV(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- Percent
- description :
- 2011 NLCD Imperviousness Percent
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- FRC_URB2D(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- fraction
- description :
- Urban fraction derived from 30 m NLCD 2011 (22 = 50%, 23 = 90%, 24 = 95%)
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- URB_PARAM(Time, z-dimension0132, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XYZ
- units :
- dimensionless
- description :
- Urban_Parameters
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[23950080 values with dtype=float32]
- LAKE_DEPTH(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- meters MSL
- description :
- Topography height
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- VAR_SSO(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- meters2 MSL
- description :
- Variance of Subgrid Scale Orography
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- OL4(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- whoknows
- description :
- something
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- OL3(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- whoknows
- description :
- something
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- OL2(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- whoknows
- description :
- something
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- OL1(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- whoknows
- description :
- something
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- OA4(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- whoknows
- description :
- something
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- OA3(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- whoknows
- description :
- something
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- OA2(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- whoknows
- description :
- something
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- OA1(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- whoknows
- description :
- something
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- VAR(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- whoknows
- description :
- something
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- CON(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- whoknows
- description :
- something
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- SNOALB(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- percent
- description :
- MODIS maximum snow albedo
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- LAI12M(Time, z-dimension0012, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XYZ
- units :
- m^2/m^2
- description :
- MODIS LAI
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[2177280 values with dtype=float32]
- GREENFRAC(Time, z-dimension0012, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XYZ
- units :
- fraction
- description :
- MODIS FPAR
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[2177280 values with dtype=float32]
- ALBEDO12M(Time, z-dimension0012, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XYZ
- units :
- percent
- description :
- Monthly MODIS surface albedo
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[2177280 values with dtype=float32]
- SCB_DOM(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- category
- description :
- Dominant category
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- SOILCBOT(Time, z-dimension0016, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XYZ
- units :
- category
- description :
- 16-category bottom-layer soil type
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[2903040 values with dtype=float32]
- SCT_DOM(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- category
- description :
- Dominant category
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- SOILCTOP(Time, z-dimension0016, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XYZ
- units :
- category
- description :
- 16-category top-layer soil type
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[2903040 values with dtype=float32]
- SOILTEMP(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- Kelvin
- description :
- Annual mean deep soil temperature
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- HGT_M(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- meters MSL
- description :
- GMTED2010 30-arc-second topography height
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- LU_INDEX(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- category
- description :
- Dominant category
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- LANDUSEF(Time, z-dimension0021, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XYZ
- units :
- category
- description :
- Noah-modified 21-category IGBP-MODIS landuse
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[3810240 values with dtype=float32]
- COSALPHA_V(Time, south_north_stag, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- none
- description :
- Cosine of rotation angle on V grid
- stagger :
- V
- sr_x :
- 1
- sr_y :
- 1
[181920 values with dtype=float32]
- SINALPHA_V(Time, south_north_stag, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- none
- description :
- Sine of rotation angle on V grid
- stagger :
- V
- sr_x :
- 1
- sr_y :
- 1
[181920 values with dtype=float32]
- COSALPHA_U(Time, south_north, west_east_stag)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- none
- description :
- Cosine of rotation angle on U grid
- stagger :
- U
- sr_x :
- 1
- sr_y :
- 1
[181818 values with dtype=float32]
- SINALPHA_U(Time, south_north, west_east_stag)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- none
- description :
- Sine of rotation angle on U grid
- stagger :
- U
- sr_x :
- 1
- sr_y :
- 1
[181818 values with dtype=float32]
- XLONG_C(Time, south_north_stag, west_east_stag)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- degrees longitude
- description :
- Longitude at grid cell corners
- stagger :
- CORNER
- sr_x :
- 1
- sr_y :
- 1
[182299 values with dtype=float32]
- XLAT_C(Time, south_north_stag, west_east_stag)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- degrees latitude
- description :
- Latitude at grid cell corners
- stagger :
- CORNER
- sr_x :
- 1
- sr_y :
- 1
[182299 values with dtype=float32]
- LANDMASK(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- none
- description :
- Landmask : 1=land, 0=water
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- COSALPHA(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- none
- description :
- Cosine of rotation angle
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- SINALPHA(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- none
- description :
- Sine of rotation angle
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- F(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- -
- description :
- Coriolis F parameter
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- E(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- -
- description :
- Coriolis E parameter
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- MAPFAC_UY(Time, south_north, west_east_stag)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- none
- description :
- Mapfactor (y-dir) on U grid
- stagger :
- U
- sr_x :
- 1
- sr_y :
- 1
[181818 values with dtype=float32]
- MAPFAC_VY(Time, south_north_stag, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- none
- description :
- Mapfactor (y-dir) on V grid
- stagger :
- V
- sr_x :
- 1
- sr_y :
- 1
[181920 values with dtype=float32]
- MAPFAC_MY(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- none
- description :
- Mapfactor (y-dir) on mass grid
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- MAPFAC_UX(Time, south_north, west_east_stag)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- none
- description :
- Mapfactor (x-dir) on U grid
- stagger :
- U
- sr_x :
- 1
- sr_y :
- 1
[181818 values with dtype=float32]
- MAPFAC_VX(Time, south_north_stag, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- none
- description :
- Mapfactor (x-dir) on V grid
- stagger :
- V
- sr_x :
- 1
- sr_y :
- 1
[181920 values with dtype=float32]
- MAPFAC_MX(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- none
- description :
- Mapfactor (x-dir) on mass grid
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- MAPFAC_U(Time, south_north, west_east_stag)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- none
- description :
- Mapfactor on U grid
- stagger :
- U
- sr_x :
- 1
- sr_y :
- 1
[181818 values with dtype=float32]
- MAPFAC_V(Time, south_north_stag, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- none
- description :
- Mapfactor on V grid
- stagger :
- V
- sr_x :
- 1
- sr_y :
- 1
[181920 values with dtype=float32]
- MAPFAC_M(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- none
- description :
- Mapfactor on mass grid
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- CLONG(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- degrees longitude
- description :
- Computational longitude on mass grid
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- CLAT(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- degrees latitude
- description :
- Computational latitude on mass grid
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- XLONG_U(Time, south_north, west_east_stag)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- degrees longitude
- description :
- Longitude on U grid
- stagger :
- U
- sr_x :
- 1
- sr_y :
- 1
[181818 values with dtype=float32]
- XLAT_U(Time, south_north, west_east_stag)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- degrees latitude
- description :
- Latitude on U grid
- stagger :
- U
- sr_x :
- 1
- sr_y :
- 1
[181818 values with dtype=float32]
- XLONG_V(Time, south_north_stag, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- degrees longitude
- description :
- Longitude on V grid
- stagger :
- V
- sr_x :
- 1
- sr_y :
- 1
[181920 values with dtype=float32]
- XLAT_V(Time, south_north_stag, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- degrees latitude
- description :
- Latitude on V grid
- stagger :
- V
- sr_x :
- 1
- sr_y :
- 1
[181920 values with dtype=float32]
- XLONG_M(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- degrees longitude
- description :
- Longitude on mass grid
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- XLAT_M(Time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- degrees latitude
- description :
- Latitude on mass grid
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
[181440 values with dtype=float32]
- TITLE :
- OUTPUT FROM METGRID V4.3
- SIMULATION_START_DATE :
- 1954-02-17_00:00:00
- WEST-EAST_GRID_DIMENSION :
- 481
- SOUTH-NORTH_GRID_DIMENSION :
- 379
- BOTTOM-TOP_GRID_DIMENSION :
- 38
- WEST-EAST_PATCH_START_UNSTAG :
- 1
- WEST-EAST_PATCH_END_UNSTAG :
- 480
- WEST-EAST_PATCH_START_STAG :
- 1
- WEST-EAST_PATCH_END_STAG :
- 481
- SOUTH-NORTH_PATCH_START_UNSTAG :
- 1
- SOUTH-NORTH_PATCH_END_UNSTAG :
- 378
- SOUTH-NORTH_PATCH_START_STAG :
- 1
- SOUTH-NORTH_PATCH_END_STAG :
- 379
- GRIDTYPE :
- C
- DX :
- 12000.0
- DY :
- 12000.0
- DYN_OPT :
- 2
- CEN_LAT :
- -20.500023
- CEN_LON :
- 145.0
- TRUELAT1 :
- -27.0
- TRUELAT2 :
- -13.0
- MOAD_CEN_LAT :
- -20.500023
- STAND_LON :
- 145.0
- POLE_LAT :
- 90.0
- POLE_LON :
- 0.0
- corner_lats :
- [-38.206017 1.402092 1.402092 -38.206017 -38.196312 1.4094925 1.4094925 -38.196312 -38.25664 1.4523087 1.4523087 -38.25664 -38.24693 1.4596939 1.4596939 -38.24693 ]
- corner_lons :
- [113.38779 120.59003 169.40997 176.61218 113.323364 120.539795 169.4602 176.67664 113.37543 120.59741 169.40259 176.62457 113.310974 120.54721 169.45282 176.68903 ]
- MAP_PROJ :
- 1
- MMINLU :
- MODIFIED_IGBP_MODIS_NOAH
- NUM_LAND_CAT :
- 21
- ISWATER :
- 17
- ISLAKE :
- 21
- ISICE :
- 15
- ISURBAN :
- 13
- ISOILWATER :
- 14
- grid_id :
- 1
- parent_id :
- 1
- i_parent_start :
- 1
- j_parent_start :
- 1
- i_parent_end :
- 481
- j_parent_end :
- 379
- parent_grid_ratio :
- 1
- sr_x :
- 1
- sr_y :
- 1
- NUM_METGRID_SOIL_LEVELS :
- 4
- FLAG_METGRID :
- 1
- FLAG_EXCLUDED_MIDDLE :
- 0
- FLAG_SOIL_LAYERS :
- 1
- FLAG_SNOW :
- 1
- FLAG_PSFC :
- 1
- FLAG_SM000007 :
- 1
- FLAG_SM007028 :
- 1
- FLAG_SM028100 :
- 1
- FLAG_SM100289 :
- 1
- FLAG_ST000007 :
- 1
- FLAG_ST007028 :
- 1
- FLAG_ST028100 :
- 1
- FLAG_ST100289 :
- 1
- FLAG_SLP :
- 1
- FLAG_SST :
- 1
- FLAG_SNOWH :
- 1
- FLAG_SOILHGT :
- 1
- FLAG_MF_XY :
- 1
- FLAG_LAI12M :
- 1
- FLAG_VAR_SSO :
- 1
- FLAG_LAKE_DEPTH :
- 1
- FLAG_URB_PARAM :
- 1
- FLAG_FRC_URB2D :
- 1
- FLAG_IMPERV :
- 1
- FLAG_CANFRA :
- 1
- FLAG_EROD :
- 1
- FLAG_CLAYFRAC :
- 1
- FLAG_SANDFRAC :
- 1
Well that wasn’t super helpful. Xarray has trouble unpacking the variables in the files, primarily in regards to the coordinates! It lumps the coordinates together with the variables, makes this difficult to work with. As mentioned previously, the data are not CF-compliant (aka it’s not neccessarily Xarray’s fault), which makes it challenging to parse correctly. This is where xwrf can help!
Investigate the Variables#
Before we read the data in using xwrf, we can take a look at which variables we would like to subset! The description attribute provides a helpful variable name, which we can take a look at using the following loop!
for var in wrf_ds:
try:
print(f'variable: {var}, description: {wrf_ds[var].description}')
except:
pass
variable: PRES, description:
variable: SOIL_LAYERS, description:
variable: SM, description:
variable: ST, description:
variable: GHT, description: Height
variable: SM100289, description: Soil moisture of 100-289 cm ground layer
variable: SM028100, description: Soil moisture of 28-100 cm ground layer
variable: SM007028, description: Soil moisture of 7-28 cm ground layer
variable: SM000007, description: Soil moisture of 0-7 cm ground layer
variable: ST100289, description: T of 100-289 cm ground layer
variable: ST028100, description: T of 28-100 cm ground layer
variable: ST007028, description: T of 7-28 cm ground layer
variable: ST000007, description: T of 0-7 cm ground layer
variable: SNOWH, description: Physical Snow Depth
variable: SNOW, description: Water Equivalent of Accumulated Snow Depth
variable: SST, description: Sea-Surface Temperature
variable: SEAICE, description: Sea-Ice Fraction
variable: SKINTEMP, description: Sea-Surface Temperature
variable: PMSL, description: Sea-level Pressure
variable: PSFC, description: Surface Pressure
variable: RH, description: Relative Humidity
variable: VV, description: V
variable: UU, description: U
variable: TT, description: Temperature
variable: SOILHGT, description: Terrain field of source analysis
variable: LANDSEA, description: Land/Sea flag
variable: OL4SS, description: orographic effective length for North-Westerly flow (small-scale)
variable: OL3SS, description: orographic effective length for South-Westerly flow (small-scale)
variable: OL2SS, description: orographic effective length for Southerly flow (small-scale)
variable: OL1SS, description: orographic effective length for Westerly flow (small-scale)
variable: OA4SS, description: orographic asymmetry in North-West direction (small-scale)
variable: OA3SS, description: orographic asymmetry in South-West direction (small-scale)
variable: OA2SS, description: orographic asymmetry in South direction (small-scale)
variable: OA1SS, description: orographic asymmetry in West direction (small-scale)
variable: VARSS, description: standard deviation of small-scale topography
variable: CONSS, description: convexity of small-scale topography
variable: OL4LS, description: orographic effective length (detrended) for North-Westerly flow
variable: OL3LS, description: orographic effective length (detrended) for South-Westerly flow
variable: OL2LS, description: orographic effective length (detrended) for Southerly flow
variable: OL1LS, description: orographic effective length (detrended) for Westerly flow
variable: OA4LS, description: orographic asymmetry (detrended) in North-West direction
variable: OA3LS, description: orographic asymmetry (detrended) in South-West direction
variable: OA2LS, description: orographic asymmetry (detrended) in South
variable: OA1LS, description: orographic asymmetry (detrended) in West direction
variable: VARLS, description: standard deviation of detrended topography
variable: CONLS, description: convexity of detrended topography
variable: IRRIGATION, description: irrigated land percentage
variable: SANDFRAC, description: Sand Fraction
variable: CLAYFRAC, description: Clay Fraction
variable: EROD, description: EROD
variable: CANFRA, description: 2011 NLCD Canopy Percent
variable: IMPERV, description: 2011 NLCD Imperviousness Percent
variable: FRC_URB2D, description: Urban fraction derived from 30 m NLCD 2011 (22 = 50%, 23 = 90%, 24 = 95%)
variable: URB_PARAM, description: Urban_Parameters
variable: LAKE_DEPTH, description: Topography height
variable: VAR_SSO, description: Variance of Subgrid Scale Orography
variable: OL4, description: something
variable: OL3, description: something
variable: OL2, description: something
variable: OL1, description: something
variable: OA4, description: something
variable: OA3, description: something
variable: OA2, description: something
variable: OA1, description: something
variable: VAR, description: something
variable: CON, description: something
variable: SNOALB, description: MODIS maximum snow albedo
variable: LAI12M, description: MODIS LAI
variable: GREENFRAC, description: MODIS FPAR
variable: ALBEDO12M, description: Monthly MODIS surface albedo
variable: SCB_DOM, description: Dominant category
variable: SOILCBOT, description: 16-category bottom-layer soil type
variable: SCT_DOM, description: Dominant category
variable: SOILCTOP, description: 16-category top-layer soil type
variable: SOILTEMP, description: Annual mean deep soil temperature
variable: HGT_M, description: GMTED2010 30-arc-second topography height
variable: LU_INDEX, description: Dominant category
variable: LANDUSEF, description: Noah-modified 21-category IGBP-MODIS landuse
variable: COSALPHA_V, description: Cosine of rotation angle on V grid
variable: SINALPHA_V, description: Sine of rotation angle on V grid
variable: COSALPHA_U, description: Cosine of rotation angle on U grid
variable: SINALPHA_U, description: Sine of rotation angle on U grid
variable: XLONG_C, description: Longitude at grid cell corners
variable: XLAT_C, description: Latitude at grid cell corners
variable: LANDMASK, description: Landmask : 1=land, 0=water
variable: COSALPHA, description: Cosine of rotation angle
variable: SINALPHA, description: Sine of rotation angle
variable: F, description: Coriolis F parameter
variable: E, description: Coriolis E parameter
variable: MAPFAC_UY, description: Mapfactor (y-dir) on U grid
variable: MAPFAC_VY, description: Mapfactor (y-dir) on V grid
variable: MAPFAC_MY, description: Mapfactor (y-dir) on mass grid
variable: MAPFAC_UX, description: Mapfactor (x-dir) on U grid
variable: MAPFAC_VX, description: Mapfactor (x-dir) on V grid
variable: MAPFAC_MX, description: Mapfactor (x-dir) on mass grid
variable: MAPFAC_U, description: Mapfactor on U grid
variable: MAPFAC_V, description: Mapfactor on V grid
variable: MAPFAC_M, description: Mapfactor on mass grid
variable: CLONG, description: Computational longitude on mass grid
variable: CLAT, description: Computational latitude on mass grid
variable: XLONG_U, description: Longitude on U grid
variable: XLAT_U, description: Latitude on U grid
variable: XLONG_V, description: Longitude on V grid
variable: XLAT_V, description: Latitude on V grid
variable: XLONG_M, description: Longitude on mass grid
variable: XLAT_M, description: Latitude on mass grid
Let’s take a look at a couple of variables - temperature and moisture!
Temperature
Relative Humidity
Read in the Dataset#
Let’s load in the data using the xwrf backend! We subset for our variables using the preprocess function below.
%%time
variables = ["PRES", "TT", "RH"]
def preprocess(ds):
return ds[variables]
ds = xr.open_mfdataset(
file_subset,
engine="xwrf",
parallel=True,
concat_dim="Time",
combine="nested",
preprocess=preprocess,
chunks={'Time': 1},
)
CPU times: user 4.94 s, sys: 1.19 s, total: 6.13 s
Wall time: 18.1 s
Investigate the Data#
ds
<xarray.Dataset>
Dimensions: (Time: 80, num_metgrid_levels: 38, south_north: 378, west_east: 480)
Coordinates:
Times (Time) datetime64[ns] dask.array<chunksize=(1,), meta=np.ndarray>
CLONG (south_north, west_east) float32 dask.array<chunksize=(378, 480), meta=np.ndarray>
CLAT (south_north, west_east) float32 dask.array<chunksize=(378, 480), meta=np.ndarray>
XLONG_M (south_north, west_east) float32 dask.array<chunksize=(378, 480), meta=np.ndarray>
XLAT_M (south_north, west_east) float32 dask.array<chunksize=(378, 480), meta=np.ndarray>
Dimensions without coordinates: Time, num_metgrid_levels, south_north, west_east
Data variables:
PRES (Time, num_metgrid_levels, south_north, west_east) float32 dask.array<chunksize=(1, 38, 378, 480), meta=np.ndarray>
TT (Time, num_metgrid_levels, south_north, west_east) float32 dask.array<chunksize=(1, 38, 378, 480), meta=np.ndarray>
RH (Time, num_metgrid_levels, south_north, west_east) float32 dask.array<chunksize=(1, 38, 378, 480), meta=np.ndarray>
Attributes: (12/73)
TITLE: OUTPUT FROM METGRID V4.3
SIMULATION_START_DATE: 2017-03-28_03:00:00
WEST-EAST_GRID_DIMENSION: 481
SOUTH-NORTH_GRID_DIMENSION: 379
BOTTOM-TOP_GRID_DIMENSION: 38
WEST-EAST_PATCH_START_UNSTAG: 1
... ...
FLAG_FRC_URB2D: 1
FLAG_IMPERV: 1
FLAG_CANFRA: 1
FLAG_EROD: 1
FLAG_CLAYFRAC: 1
FLAG_SANDFRAC: 1- Time: 80
- num_metgrid_levels: 38
- south_north: 378
- west_east: 480
- Times(Time)datetime64[ns]dask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 640 B 8 B Shape (80,) (1,) Count 240 Tasks 80 Chunks Type datetime64[ns] numpy.ndarray - CLONG(south_north, west_east)float32dask.array<chunksize=(378, 480), meta=np.ndarray>
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- degrees longitude
- description :
- Computational longitude on mass grid
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
Array Chunk Bytes 708.75 kiB 708.75 kiB Shape (378, 480) (378, 480) Count 395 Tasks 1 Chunks Type float32 numpy.ndarray - CLAT(south_north, west_east)float32dask.array<chunksize=(378, 480), meta=np.ndarray>
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- degrees latitude
- description :
- Computational latitude on mass grid
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
Array Chunk Bytes 708.75 kiB 708.75 kiB Shape (378, 480) (378, 480) Count 395 Tasks 1 Chunks Type float32 numpy.ndarray - XLONG_M(south_north, west_east)float32dask.array<chunksize=(378, 480), meta=np.ndarray>
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- degrees longitude
- description :
- Longitude on mass grid
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
Array Chunk Bytes 708.75 kiB 708.75 kiB Shape (378, 480) (378, 480) Count 395 Tasks 1 Chunks Type float32 numpy.ndarray - XLAT_M(south_north, west_east)float32dask.array<chunksize=(378, 480), meta=np.ndarray>
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- degrees latitude
- description :
- Latitude on mass grid
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
Array Chunk Bytes 708.75 kiB 708.75 kiB Shape (378, 480) (378, 480) Count 395 Tasks 1 Chunks Type float32 numpy.ndarray
- PRES(Time, num_metgrid_levels, south_north, west_east)float32dask.array<chunksize=(1, 38, 378, 480), meta=np.ndarray>
- FieldType :
- 104
- MemoryOrder :
- XYZ
- units :
- description :
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
Array Chunk Bytes 2.05 GiB 26.30 MiB Shape (80, 38, 378, 480) (1, 38, 378, 480) Count 240 Tasks 80 Chunks Type float32 numpy.ndarray - TT(Time, num_metgrid_levels, south_north, west_east)float32dask.array<chunksize=(1, 38, 378, 480), meta=np.ndarray>
- FieldType :
- 104
- MemoryOrder :
- XYZ
- units :
- K
- description :
- Temperature
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
Array Chunk Bytes 2.05 GiB 26.30 MiB Shape (80, 38, 378, 480) (1, 38, 378, 480) Count 240 Tasks 80 Chunks Type float32 numpy.ndarray - RH(Time, num_metgrid_levels, south_north, west_east)float32dask.array<chunksize=(1, 38, 378, 480), meta=np.ndarray>
- FieldType :
- 104
- MemoryOrder :
- XYZ
- units :
- %
- description :
- Relative Humidity
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
Array Chunk Bytes 2.05 GiB 26.30 MiB Shape (80, 38, 378, 480) (1, 38, 378, 480) Count 240 Tasks 80 Chunks Type float32 numpy.ndarray
- TITLE :
- OUTPUT FROM METGRID V4.3
- SIMULATION_START_DATE :
- 2017-03-28_03:00:00
- WEST-EAST_GRID_DIMENSION :
- 481
- SOUTH-NORTH_GRID_DIMENSION :
- 379
- BOTTOM-TOP_GRID_DIMENSION :
- 38
- WEST-EAST_PATCH_START_UNSTAG :
- 1
- WEST-EAST_PATCH_END_UNSTAG :
- 480
- WEST-EAST_PATCH_START_STAG :
- 1
- WEST-EAST_PATCH_END_STAG :
- 481
- SOUTH-NORTH_PATCH_START_UNSTAG :
- 1
- SOUTH-NORTH_PATCH_END_UNSTAG :
- 378
- SOUTH-NORTH_PATCH_START_STAG :
- 1
- SOUTH-NORTH_PATCH_END_STAG :
- 379
- GRIDTYPE :
- C
- DX :
- 12000.0
- DY :
- 12000.0
- DYN_OPT :
- 2
- CEN_LAT :
- -20.500023
- CEN_LON :
- 145.0
- TRUELAT1 :
- -27.0
- TRUELAT2 :
- -13.0
- MOAD_CEN_LAT :
- -20.500023
- STAND_LON :
- 145.0
- POLE_LAT :
- 90.0
- POLE_LON :
- 0.0
- corner_lats :
- [-38.206017 1.402092 1.402092 -38.206017 -38.196312 1.4094925 1.4094925 -38.196312 -38.25664 1.4523087 1.4523087 -38.25664 -38.24693 1.4596939 1.4596939 -38.24693 ]
- corner_lons :
- [113.38779 120.59003 169.40997 176.61218 113.323364 120.539795 169.4602 176.67664 113.37543 120.59741 169.40259 176.62457 113.310974 120.54721 169.45282 176.68903 ]
- MAP_PROJ :
- 1
- MMINLU :
- MODIFIED_IGBP_MODIS_NOAH
- NUM_LAND_CAT :
- 21
- ISWATER :
- 17
- ISLAKE :
- 21
- ISICE :
- 15
- ISURBAN :
- 13
- ISOILWATER :
- 14
- grid_id :
- 1
- parent_id :
- 1
- i_parent_start :
- 1
- j_parent_start :
- 1
- i_parent_end :
- 481
- j_parent_end :
- 379
- parent_grid_ratio :
- 1
- sr_x :
- 1
- sr_y :
- 1
- NUM_METGRID_SOIL_LEVELS :
- 4
- FLAG_METGRID :
- 1
- FLAG_EXCLUDED_MIDDLE :
- 0
- FLAG_SOIL_LAYERS :
- 1
- FLAG_SNOW :
- 1
- FLAG_PSFC :
- 1
- FLAG_SM000007 :
- 1
- FLAG_SM007028 :
- 1
- FLAG_SM028100 :
- 1
- FLAG_SM100289 :
- 1
- FLAG_ST000007 :
- 1
- FLAG_ST007028 :
- 1
- FLAG_ST028100 :
- 1
- FLAG_ST100289 :
- 1
- FLAG_SLP :
- 1
- FLAG_SST :
- 1
- FLAG_SNOWH :
- 1
- FLAG_SOILHGT :
- 1
- FLAG_MF_XY :
- 1
- FLAG_LAI12M :
- 1
- FLAG_VAR_SSO :
- 1
- FLAG_LAKE_DEPTH :
- 1
- FLAG_URB_PARAM :
- 1
- FLAG_FRC_URB2D :
- 1
- FLAG_IMPERV :
- 1
- FLAG_CANFRA :
- 1
- FLAG_EROD :
- 1
- FLAG_CLAYFRAC :
- 1
- FLAG_SANDFRAC :
- 1
Investigate the Vertical Levels#
Each of the variables as a vertical dimension of num_metgrid_levels, indicating the index (from 0 to 37) of the vertical level. This value isn’t super helpful; what would be helpful is a vertical dimension corresponding to the pressure level.
If we look at a single grid point, you will notice that the bottom level (num_metgrid_levels=0) is the bottom pressure level. These change across time!
ds.PRES.isel(num_metgrid_levels=0, south_north=0, west_east=0).values
array([101763.984, 101914.88 , 102096.47 , 102343.336, 102487.016,
102629.72 , 102679.38 , 102858.94 , 102934.56 , 102820.25 ,
102769.125, 102777.82 , 102698.086, 102618.266, 102534.625,
102537.375, 102484.625, 102379.58 , 102359.77 , 102492.22 ,
102578.125, 102686.39 , 102788.92 , 102977.1 , 103049.72 ,
103027.08 , 103026. , 103122.66 , 103085.516, 103014.25 ,
102992.19 , 103042.41 , 102982.3 , 102737.12 , 102663.31 ,
102644.55 , 102549.31 , 102404.51 , 102274.69 , 102241.29 ,
102118.38 , 102049.37 , 102067.3 , 102203.125, 102274.09 ,
102373.07 , 102488.93 , 102695.16 , 102835.19 , 102877.78 ,
102894.4 , 103038.38 , 103036.32 , 103004.57 , 102952.06 ,
103012.39 , 102951.68 , 102752. , 102648.27 , 102613.55 ,
102496.516, 102351.45 , 102215.32 , 102282.81 , 102211.734,
102048.64 , 101986.07 , 102015.19 , 102028.39 , 101949.195,
101925.83 , 102046.484, 102018.53 , 101890.97 , 102007.336,
102205.65 , 102318.29 , 102340.164, 102362.14 , 102449.94 ],
dtype=float32)
If we move up one more level, you will see that we now have an a pressure level that is consistent across time; which is true across the rest of these vertical levels!
ds.PRES.isel(num_metgrid_levels=1, south_north=0, west_east=0).values
array([100000., 100000., 100000., 100000., 100000., 100000., 100000.,
100000., 100000., 100000., 100000., 100000., 100000., 100000.,
100000., 100000., 100000., 100000., 100000., 100000., 100000.,
100000., 100000., 100000., 100000., 100000., 100000., 100000.,
100000., 100000., 100000., 100000., 100000., 100000., 100000.,
100000., 100000., 100000., 100000., 100000., 100000., 100000.,
100000., 100000., 100000., 100000., 100000., 100000., 100000.,
100000., 100000., 100000., 100000., 100000., 100000., 100000.,
100000., 100000., 100000., 100000., 100000., 100000., 100000.,
100000., 100000., 100000., 100000., 100000., 100000., 100000.,
100000., 100000., 100000., 100000., 100000., 100000., 100000.,
100000., 100000., 100000.], dtype=float32)
Let’s subset the data between 1000 to 300 hPa!
ds = ds.isel(num_metgrid_levels=range(1, 21))
/glade/work/mgrover/miniconda3/envs/cesm-collections-dev/lib/python3.9/site-packages/xarray/core/indexing.py:1226: PerformanceWarning: Slicing is producing a large chunk. To accept the large
chunk and silence this warning, set the option
>>> with dask.config.set(**{'array.slicing.split_large_chunks': False}):
... array[indexer]
To avoid creating the large chunks, set the option
>>> with dask.config.set(**{'array.slicing.split_large_chunks': True}):
... array[indexer]
return self.array[key]
Next, we can rename the vertical levels to plev which stands for pressure levels!
ds_to_plot = ds.rename({'num_metgrid_levels': 'plev'})
ds_to_plot
<xarray.Dataset>
Dimensions: (Time: 80, plev: 20, south_north: 378, west_east: 480)
Coordinates:
Times (Time) datetime64[ns] dask.array<chunksize=(1,), meta=np.ndarray>
CLONG (south_north, west_east) float32 dask.array<chunksize=(378, 480), meta=np.ndarray>
CLAT (south_north, west_east) float32 dask.array<chunksize=(378, 480), meta=np.ndarray>
XLONG_M (south_north, west_east) float32 dask.array<chunksize=(378, 480), meta=np.ndarray>
XLAT_M (south_north, west_east) float32 dask.array<chunksize=(378, 480), meta=np.ndarray>
Dimensions without coordinates: Time, plev, south_north, west_east
Data variables:
PRES (Time, plev, south_north, west_east) float32 dask.array<chunksize=(1, 20, 378, 480), meta=np.ndarray>
TT (Time, plev, south_north, west_east) float32 dask.array<chunksize=(1, 20, 378, 480), meta=np.ndarray>
RH (Time, plev, south_north, west_east) float32 dask.array<chunksize=(1, 20, 378, 480), meta=np.ndarray>
Attributes: (12/73)
TITLE: OUTPUT FROM METGRID V4.3
SIMULATION_START_DATE: 2017-03-28_03:00:00
WEST-EAST_GRID_DIMENSION: 481
SOUTH-NORTH_GRID_DIMENSION: 379
BOTTOM-TOP_GRID_DIMENSION: 38
WEST-EAST_PATCH_START_UNSTAG: 1
... ...
FLAG_FRC_URB2D: 1
FLAG_IMPERV: 1
FLAG_CANFRA: 1
FLAG_EROD: 1
FLAG_CLAYFRAC: 1
FLAG_SANDFRAC: 1- Time: 80
- plev: 20
- south_north: 378
- west_east: 480
- Times(Time)datetime64[ns]dask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 640 B 8 B Shape (80,) (1,) Count 240 Tasks 80 Chunks Type datetime64[ns] numpy.ndarray - CLONG(south_north, west_east)float32dask.array<chunksize=(378, 480), meta=np.ndarray>
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- degrees longitude
- description :
- Computational longitude on mass grid
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
Array Chunk Bytes 708.75 kiB 708.75 kiB Shape (378, 480) (378, 480) Count 395 Tasks 1 Chunks Type float32 numpy.ndarray - CLAT(south_north, west_east)float32dask.array<chunksize=(378, 480), meta=np.ndarray>
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- degrees latitude
- description :
- Computational latitude on mass grid
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
Array Chunk Bytes 708.75 kiB 708.75 kiB Shape (378, 480) (378, 480) Count 395 Tasks 1 Chunks Type float32 numpy.ndarray - XLONG_M(south_north, west_east)float32dask.array<chunksize=(378, 480), meta=np.ndarray>
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- degrees longitude
- description :
- Longitude on mass grid
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
Array Chunk Bytes 708.75 kiB 708.75 kiB Shape (378, 480) (378, 480) Count 395 Tasks 1 Chunks Type float32 numpy.ndarray - XLAT_M(south_north, west_east)float32dask.array<chunksize=(378, 480), meta=np.ndarray>
- FieldType :
- 104
- MemoryOrder :
- XY
- units :
- degrees latitude
- description :
- Latitude on mass grid
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
Array Chunk Bytes 708.75 kiB 708.75 kiB Shape (378, 480) (378, 480) Count 395 Tasks 1 Chunks Type float32 numpy.ndarray
- PRES(Time, plev, south_north, west_east)float32dask.array<chunksize=(1, 20, 378, 480), meta=np.ndarray>
- FieldType :
- 104
- MemoryOrder :
- XYZ
- units :
- description :
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
Array Chunk Bytes 1.08 GiB 13.84 MiB Shape (80, 20, 378, 480) (1, 20, 378, 480) Count 320 Tasks 80 Chunks Type float32 numpy.ndarray - TT(Time, plev, south_north, west_east)float32dask.array<chunksize=(1, 20, 378, 480), meta=np.ndarray>
- FieldType :
- 104
- MemoryOrder :
- XYZ
- units :
- K
- description :
- Temperature
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
Array Chunk Bytes 1.08 GiB 13.84 MiB Shape (80, 20, 378, 480) (1, 20, 378, 480) Count 320 Tasks 80 Chunks Type float32 numpy.ndarray - RH(Time, plev, south_north, west_east)float32dask.array<chunksize=(1, 20, 378, 480), meta=np.ndarray>
- FieldType :
- 104
- MemoryOrder :
- XYZ
- units :
- %
- description :
- Relative Humidity
- stagger :
- M
- sr_x :
- 1
- sr_y :
- 1
Array Chunk Bytes 1.08 GiB 13.84 MiB Shape (80, 20, 378, 480) (1, 20, 378, 480) Count 320 Tasks 80 Chunks Type float32 numpy.ndarray
- TITLE :
- OUTPUT FROM METGRID V4.3
- SIMULATION_START_DATE :
- 2017-03-28_03:00:00
- WEST-EAST_GRID_DIMENSION :
- 481
- SOUTH-NORTH_GRID_DIMENSION :
- 379
- BOTTOM-TOP_GRID_DIMENSION :
- 38
- WEST-EAST_PATCH_START_UNSTAG :
- 1
- WEST-EAST_PATCH_END_UNSTAG :
- 480
- WEST-EAST_PATCH_START_STAG :
- 1
- WEST-EAST_PATCH_END_STAG :
- 481
- SOUTH-NORTH_PATCH_START_UNSTAG :
- 1
- SOUTH-NORTH_PATCH_END_UNSTAG :
- 378
- SOUTH-NORTH_PATCH_START_STAG :
- 1
- SOUTH-NORTH_PATCH_END_STAG :
- 379
- GRIDTYPE :
- C
- DX :
- 12000.0
- DY :
- 12000.0
- DYN_OPT :
- 2
- CEN_LAT :
- -20.500023
- CEN_LON :
- 145.0
- TRUELAT1 :
- -27.0
- TRUELAT2 :
- -13.0
- MOAD_CEN_LAT :
- -20.500023
- STAND_LON :
- 145.0
- POLE_LAT :
- 90.0
- POLE_LON :
- 0.0
- corner_lats :
- [-38.206017 1.402092 1.402092 -38.206017 -38.196312 1.4094925 1.4094925 -38.196312 -38.25664 1.4523087 1.4523087 -38.25664 -38.24693 1.4596939 1.4596939 -38.24693 ]
- corner_lons :
- [113.38779 120.59003 169.40997 176.61218 113.323364 120.539795 169.4602 176.67664 113.37543 120.59741 169.40259 176.62457 113.310974 120.54721 169.45282 176.68903 ]
- MAP_PROJ :
- 1
- MMINLU :
- MODIFIED_IGBP_MODIS_NOAH
- NUM_LAND_CAT :
- 21
- ISWATER :
- 17
- ISLAKE :
- 21
- ISICE :
- 15
- ISURBAN :
- 13
- ISOILWATER :
- 14
- grid_id :
- 1
- parent_id :
- 1
- i_parent_start :
- 1
- j_parent_start :
- 1
- i_parent_end :
- 481
- j_parent_end :
- 379
- parent_grid_ratio :
- 1
- sr_x :
- 1
- sr_y :
- 1
- NUM_METGRID_SOIL_LEVELS :
- 4
- FLAG_METGRID :
- 1
- FLAG_EXCLUDED_MIDDLE :
- 0
- FLAG_SOIL_LAYERS :
- 1
- FLAG_SNOW :
- 1
- FLAG_PSFC :
- 1
- FLAG_SM000007 :
- 1
- FLAG_SM007028 :
- 1
- FLAG_SM028100 :
- 1
- FLAG_SM100289 :
- 1
- FLAG_ST000007 :
- 1
- FLAG_ST007028 :
- 1
- FLAG_ST028100 :
- 1
- FLAG_ST100289 :
- 1
- FLAG_SLP :
- 1
- FLAG_SST :
- 1
- FLAG_SNOWH :
- 1
- FLAG_SOILHGT :
- 1
- FLAG_MF_XY :
- 1
- FLAG_LAI12M :
- 1
- FLAG_VAR_SSO :
- 1
- FLAG_LAKE_DEPTH :
- 1
- FLAG_URB_PARAM :
- 1
- FLAG_FRC_URB2D :
- 1
- FLAG_IMPERV :
- 1
- FLAG_CANFRA :
- 1
- FLAG_EROD :
- 1
- FLAG_CLAYFRAC :
- 1
- FLAG_SANDFRAC :
- 1
plevs = ds_to_plot.PRES.isel(south_north=0, west_east=0).values[0, :]
ds_to_plot['plev'] = plevs / 100
ds_to_plot['plev'].attrs['units'] = 'hPa'
Modify the Time values as well…#
The Time dimension has a similar issue here where the values are integers, rather than usable time values. We can modify this by setting Time to be equal to the time values Times
ds_to_plot['Time'] = ds.Times
ds_to_plot.Time
<xarray.DataArray 'Time' (Time: 80)>
array(['2017-03-28T03:00:00.000000000', '2017-03-28T06:00:00.000000000',
'2017-03-28T09:00:00.000000000', '2017-03-28T12:00:00.000000000',
'2017-03-28T15:00:00.000000000', '2017-03-28T18:00:00.000000000',
'2017-03-28T21:00:00.000000000', '2017-03-29T00:00:00.000000000',
'2017-03-29T03:00:00.000000000', '2017-03-29T06:00:00.000000000',
'2017-03-29T09:00:00.000000000', '2017-03-29T12:00:00.000000000',
'2017-03-29T15:00:00.000000000', '2017-03-29T18:00:00.000000000',
'2017-03-29T21:00:00.000000000', '2017-03-30T00:00:00.000000000',
'2017-03-30T03:00:00.000000000', '2017-03-30T06:00:00.000000000',
'2017-03-30T09:00:00.000000000', '2017-03-30T12:00:00.000000000',
'2017-03-30T15:00:00.000000000', '2017-03-30T18:00:00.000000000',
'2017-03-30T21:00:00.000000000', '2017-03-31T00:00:00.000000000',
'2017-03-31T03:00:00.000000000', '2017-03-31T06:00:00.000000000',
'2017-03-31T09:00:00.000000000', '2017-03-31T12:00:00.000000000',
'2017-03-31T15:00:00.000000000', '2017-03-31T18:00:00.000000000',
'2017-03-31T21:00:00.000000000', '2017-04-01T00:00:00.000000000',
'2017-04-01T03:00:00.000000000', '2017-04-01T06:00:00.000000000',
'2017-04-01T09:00:00.000000000', '2017-04-01T12:00:00.000000000',
'2017-04-01T15:00:00.000000000', '2017-04-01T18:00:00.000000000',
'2017-04-01T21:00:00.000000000', '2017-04-02T00:00:00.000000000',
'2017-04-02T03:00:00.000000000', '2017-04-02T06:00:00.000000000',
'2017-04-02T09:00:00.000000000', '2017-04-02T12:00:00.000000000',
'2017-04-02T15:00:00.000000000', '2017-04-02T18:00:00.000000000',
'2017-04-02T21:00:00.000000000', '2017-04-03T00:00:00.000000000',
'2017-04-03T03:00:00.000000000', '2017-04-03T06:00:00.000000000',
'2017-04-03T09:00:00.000000000', '2017-04-03T12:00:00.000000000',
'2017-04-03T15:00:00.000000000', '2017-04-03T18:00:00.000000000',
'2017-04-03T21:00:00.000000000', '2017-04-04T00:00:00.000000000',
'2017-04-04T03:00:00.000000000', '2017-04-04T06:00:00.000000000',
'2017-04-04T09:00:00.000000000', '2017-04-04T12:00:00.000000000',
'2017-04-04T15:00:00.000000000', '2017-04-04T18:00:00.000000000',
'2017-04-04T21:00:00.000000000', '2017-04-05T00:00:00.000000000',
'2017-04-05T03:00:00.000000000', '2017-04-05T06:00:00.000000000',
'2017-04-05T09:00:00.000000000', '2017-04-05T12:00:00.000000000',
'2017-04-05T15:00:00.000000000', '2017-04-05T18:00:00.000000000',
'2017-04-05T21:00:00.000000000', '2017-04-06T00:00:00.000000000',
'2017-04-06T03:00:00.000000000', '2017-04-06T06:00:00.000000000',
'2017-04-06T09:00:00.000000000', '2017-04-06T12:00:00.000000000',
'2017-04-06T15:00:00.000000000', '2017-04-06T18:00:00.000000000',
'2017-04-06T21:00:00.000000000', '2017-04-07T00:00:00.000000000'],
dtype='datetime64[ns]')
Coordinates:
Times (Time) datetime64[ns] dask.array<chunksize=(1,), meta=np.ndarray>
* Time (Time) datetime64[ns] 2017-03-28T03:00:00 ... 2017-04-07- Time: 80
- 2017-03-28T03:00:00 2017-03-28T06:00:00 ... 2017-04-07
array(['2017-03-28T03:00:00.000000000', '2017-03-28T06:00:00.000000000', '2017-03-28T09:00:00.000000000', '2017-03-28T12:00:00.000000000', '2017-03-28T15:00:00.000000000', '2017-03-28T18:00:00.000000000', '2017-03-28T21:00:00.000000000', '2017-03-29T00:00:00.000000000', '2017-03-29T03:00:00.000000000', '2017-03-29T06:00:00.000000000', '2017-03-29T09:00:00.000000000', '2017-03-29T12:00:00.000000000', '2017-03-29T15:00:00.000000000', '2017-03-29T18:00:00.000000000', '2017-03-29T21:00:00.000000000', '2017-03-30T00:00:00.000000000', '2017-03-30T03:00:00.000000000', '2017-03-30T06:00:00.000000000', '2017-03-30T09:00:00.000000000', '2017-03-30T12:00:00.000000000', '2017-03-30T15:00:00.000000000', '2017-03-30T18:00:00.000000000', '2017-03-30T21:00:00.000000000', '2017-03-31T00:00:00.000000000', '2017-03-31T03:00:00.000000000', '2017-03-31T06:00:00.000000000', '2017-03-31T09:00:00.000000000', '2017-03-31T12:00:00.000000000', '2017-03-31T15:00:00.000000000', '2017-03-31T18:00:00.000000000', '2017-03-31T21:00:00.000000000', '2017-04-01T00:00:00.000000000', '2017-04-01T03:00:00.000000000', '2017-04-01T06:00:00.000000000', '2017-04-01T09:00:00.000000000', '2017-04-01T12:00:00.000000000', '2017-04-01T15:00:00.000000000', '2017-04-01T18:00:00.000000000', '2017-04-01T21:00:00.000000000', '2017-04-02T00:00:00.000000000', '2017-04-02T03:00:00.000000000', '2017-04-02T06:00:00.000000000', '2017-04-02T09:00:00.000000000', '2017-04-02T12:00:00.000000000', '2017-04-02T15:00:00.000000000', '2017-04-02T18:00:00.000000000', '2017-04-02T21:00:00.000000000', '2017-04-03T00:00:00.000000000', '2017-04-03T03:00:00.000000000', '2017-04-03T06:00:00.000000000', '2017-04-03T09:00:00.000000000', '2017-04-03T12:00:00.000000000', '2017-04-03T15:00:00.000000000', '2017-04-03T18:00:00.000000000', '2017-04-03T21:00:00.000000000', '2017-04-04T00:00:00.000000000', '2017-04-04T03:00:00.000000000', '2017-04-04T06:00:00.000000000', '2017-04-04T09:00:00.000000000', '2017-04-04T12:00:00.000000000', '2017-04-04T15:00:00.000000000', '2017-04-04T18:00:00.000000000', '2017-04-04T21:00:00.000000000', '2017-04-05T00:00:00.000000000', '2017-04-05T03:00:00.000000000', '2017-04-05T06:00:00.000000000', '2017-04-05T09:00:00.000000000', '2017-04-05T12:00:00.000000000', '2017-04-05T15:00:00.000000000', '2017-04-05T18:00:00.000000000', '2017-04-05T21:00:00.000000000', '2017-04-06T00:00:00.000000000', '2017-04-06T03:00:00.000000000', '2017-04-06T06:00:00.000000000', '2017-04-06T09:00:00.000000000', '2017-04-06T12:00:00.000000000', '2017-04-06T15:00:00.000000000', '2017-04-06T18:00:00.000000000', '2017-04-06T21:00:00.000000000', '2017-04-07T00:00:00.000000000'], dtype='datetime64[ns]') - Times(Time)datetime64[ns]dask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 640 B 8 B Shape (80,) (1,) Count 240 Tasks 80 Chunks Type datetime64[ns] numpy.ndarray - Time(Time)datetime64[ns]2017-03-28T03:00:00 ... 2017-04-07
array(['2017-03-28T03:00:00.000000000', '2017-03-28T06:00:00.000000000', '2017-03-28T09:00:00.000000000', '2017-03-28T12:00:00.000000000', '2017-03-28T15:00:00.000000000', '2017-03-28T18:00:00.000000000', '2017-03-28T21:00:00.000000000', '2017-03-29T00:00:00.000000000', '2017-03-29T03:00:00.000000000', '2017-03-29T06:00:00.000000000', '2017-03-29T09:00:00.000000000', '2017-03-29T12:00:00.000000000', '2017-03-29T15:00:00.000000000', '2017-03-29T18:00:00.000000000', '2017-03-29T21:00:00.000000000', '2017-03-30T00:00:00.000000000', '2017-03-30T03:00:00.000000000', '2017-03-30T06:00:00.000000000', '2017-03-30T09:00:00.000000000', '2017-03-30T12:00:00.000000000', '2017-03-30T15:00:00.000000000', '2017-03-30T18:00:00.000000000', '2017-03-30T21:00:00.000000000', '2017-03-31T00:00:00.000000000', '2017-03-31T03:00:00.000000000', '2017-03-31T06:00:00.000000000', '2017-03-31T09:00:00.000000000', '2017-03-31T12:00:00.000000000', '2017-03-31T15:00:00.000000000', '2017-03-31T18:00:00.000000000', '2017-03-31T21:00:00.000000000', '2017-04-01T00:00:00.000000000', '2017-04-01T03:00:00.000000000', '2017-04-01T06:00:00.000000000', '2017-04-01T09:00:00.000000000', '2017-04-01T12:00:00.000000000', '2017-04-01T15:00:00.000000000', '2017-04-01T18:00:00.000000000', '2017-04-01T21:00:00.000000000', '2017-04-02T00:00:00.000000000', '2017-04-02T03:00:00.000000000', '2017-04-02T06:00:00.000000000', '2017-04-02T09:00:00.000000000', '2017-04-02T12:00:00.000000000', '2017-04-02T15:00:00.000000000', '2017-04-02T18:00:00.000000000', '2017-04-02T21:00:00.000000000', '2017-04-03T00:00:00.000000000', '2017-04-03T03:00:00.000000000', '2017-04-03T06:00:00.000000000', '2017-04-03T09:00:00.000000000', '2017-04-03T12:00:00.000000000', '2017-04-03T15:00:00.000000000', '2017-04-03T18:00:00.000000000', '2017-04-03T21:00:00.000000000', '2017-04-04T00:00:00.000000000', '2017-04-04T03:00:00.000000000', '2017-04-04T06:00:00.000000000', '2017-04-04T09:00:00.000000000', '2017-04-04T12:00:00.000000000', '2017-04-04T15:00:00.000000000', '2017-04-04T18:00:00.000000000', '2017-04-04T21:00:00.000000000', '2017-04-05T00:00:00.000000000', '2017-04-05T03:00:00.000000000', '2017-04-05T06:00:00.000000000', '2017-04-05T09:00:00.000000000', '2017-04-05T12:00:00.000000000', '2017-04-05T15:00:00.000000000', '2017-04-05T18:00:00.000000000', '2017-04-05T21:00:00.000000000', '2017-04-06T00:00:00.000000000', '2017-04-06T03:00:00.000000000', '2017-04-06T06:00:00.000000000', '2017-04-06T09:00:00.000000000', '2017-04-06T12:00:00.000000000', '2017-04-06T15:00:00.000000000', '2017-04-06T18:00:00.000000000', '2017-04-06T21:00:00.000000000', '2017-04-07T00:00:00.000000000'], dtype='datetime64[ns]')
Plot the Output using hvPlot#
We can use hvPlot here to visualize the output. We use quadmesh to plot, which is able to use the coordinate information. Since we are not working with velocity variables, we use XLONG_M and XLAT_M for the longitude and latitude.
We also specify we only want our variables temperature (TT) and relative humidity (RH), grouping by the time (Time) and pressure level (plev)
plot = ds_to_plot.hvplot.quadmesh(
x='XLONG_M',
y='XLAT_M',
z=['TT', 'RH'],
groupby=['Time', 'plev'],
widget_location='bottom',
rasterize=True,
coastline=True,
cmap='inferno',
)
plot
Conclusions#
Within this post, we covered how to read in WRF data using a new experimental package, xwrf! We also included some example pre-processing, and a method of plotting interactive visualizations using the WRF data! We hope this helpful within your analysis, and look forward on making more progress on these examples in the future, especially as it relates to calculations utilizing Dask!