Analyzing and visualizing CAM-SE output in Python#
Overview#
We demonstrate a variety of options for analyzing and visualizing output from the Community Atmosphere Model (CAM) with the spectral element (SE) grid in Python. This notebook was developed for the ESDS Collaborative Work Time on Unstructured Grids, which took place on April 17, 2023. A recap of the related CAM-SE discussion can be found here.
Contents#
Imports#
%load_ext watermark
import distributed
import ncar_jobqueue
import numpy as np
import opt_einsum
import scipy as sp
import xarray as xr
import xesmf
%watermark -iv
sys : 3.10.10 | packaged by conda-forge | (main, Mar 24 2023, 20:08:06) [GCC 11.3.0]
xesmf : 0.7.1
distributed : 2023.4.1
json : 2.0.9
ncar_jobqueue: 2021.4.14
xarray : 2023.3.0
numpy : 1.23.5
scipy : 1.10.1
opt_einsum : 3.3.0
Spin up a Cluster#
cluster = ncar_jobqueue.NCARCluster()
cluster
PBSCluster
3d08bca5
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kdagon/proxy/8787/status | Workers: 0 |
| Total threads: 0 | Total memory: 0 B |
Scheduler Info
Scheduler
Scheduler-9a2e31b9-bbf4-4978-8e18-8c1af8bb535a
| Comm: tcp://10.12.206.45:39748 | Workers: 0 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kdagon/proxy/8787/status | Total threads: 0 |
| Started: Just now | Total memory: 0 B |
Workers
Distributed client with adaptive scaling#
client = distributed.Client(cluster)
cluster.adapt(minimum_jobs=1, maximum_jobs=4);
client
Client
Client-b37f404b-50c0-11ee-8170-3cecef19f78e
| Connection method: Cluster object | Cluster type: dask_jobqueue.PBSCluster |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kdagon/proxy/8787/status |
Cluster Info
PBSCluster
3d08bca5
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kdagon/proxy/8787/status | Workers: 0 |
| Total threads: 0 | Total memory: 0 B |
Scheduler Info
Scheduler
Scheduler-9a2e31b9-bbf4-4978-8e18-8c1af8bb535a
| Comm: tcp://10.12.206.45:39748 | Workers: 0 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kdagon/proxy/8787/status | Total threads: 0 |
| Started: Just now | Total memory: 0 B |
Workers
Set CAM-SE output location and map file#
Here we are using 3-hourly surface temperature (TS) output from a historical simulation using the ne120 grid (0.25deg resolution).
data_path = '/glade/campaign/cgd/ccr/jet/nanr_forKatie/3hrly/b.e13.B20TRC5CN.ne120_g16.003/'
data_file = 'b.e13.B20TRC5CN.ne120_g16.003.cam.h4.TS.2000010100Z-2000123121Z.nc' # TS file is 3-hourly avg for year 2000
The map file is a set of weights that is used to remap the CAM-SE unstructured output to a regular latitude by longitude grid. This particular map file uses bilinear interpolation.
ESMF_RegridWeightGen is a tool that can be used to generate map files, and this ESMPy tutorial has additional information on creating these files.
map_path = '/glade/campaign/cgd/amp/jet/ClimateNet/data_processing/maps/'
map_file = 'map_ne120_to_0.23x0.31_bilinear.nc'
1. Regrid CAM-SE output using map file#
This method aims to produce a Python-based alternative to the ncremap tool from the NCO package. The benefits of this method could include remapping within a Python script or notebook, for example to visualize unstructured data without having to write a netCDF file, remap, and read it back in. See related discussion from this issue in the UXarray github repository, and this example notebook with visualization examples from the UXarray documentation.
Read in map file (weights)#
weights = xr.open_dataset(map_path + map_file)
weights
<xarray.Dataset>
Dimensions: (src_grid_rank: 1, dst_grid_rank: 2, n_a: 777602,
n_b: 884736, nv_a: 3, nv_b: 4, n_s: 2654208)
Dimensions without coordinates: src_grid_rank, dst_grid_rank, n_a, n_b, nv_a,
nv_b, n_s
Data variables: (12/19)
src_grid_dims (src_grid_rank) int32 ...
dst_grid_dims (dst_grid_rank) int32 ...
yc_a (n_a) float64 ...
yc_b (n_b) float64 ...
xc_a (n_a) float64 ...
xc_b (n_b) float64 ...
... ...
area_b (n_b) float64 ...
frac_a (n_a) float64 ...
frac_b (n_b) float64 ...
col (n_s) int32 ...
row (n_s) int32 ...
S (n_s) float64 ...
Attributes:
title: ESMF Offline Regridding Weight Generator
normalization: destarea
map_method: Bilinear remapping
ESMF_regrid_method: Bilinear
conventions: NCAR-CSM
domain_a: /glade/p/cgd/ccr/people/nanr/mapfiles/ne120.nc
domain_b: /glade/p/cgd/ccr/people/nanr/mapfiles/0.23x0.31.nc
grid_file_src: /glade/p/cgd/ccr/people/nanr/mapfiles/ne120.nc
grid_file_dst: /glade/p/cgd/ccr/people/nanr/mapfiles/0.23x0.31.nc
CVS_revision: 6.3.0rMake source / destination grids#
srclat = weights['yc_a']
srclon = weights['xc_a']
dstlat = weights['yc_b']
dstlon = weights['xc_b']
src_grid_dims = weights['src_grid_dims'].values
dst_grid_dims = weights['dst_grid_dims'].values
print(f"Src grid dims: {src_grid_dims}, dst grid dims: {dst_grid_dims}")
n_a = weights['n_a'].size # col dimension
n_b = weights['n_b'].size # row dimension
n_s = weights['n_s'].size # nnz dimension
print(f"Map contains {n_b} rows, {n_a} cols and {n_s} nnz values")
rows = weights['row'][:] - 1 # row indices (1-based)
cols = weights['col'][:] - 1 # col indices (1-based)
nnzvals = weights['S'][:] # nnz map values
Src grid dims: [777602], dst grid dims: [1152 768]
Map contains 884736 rows, 777602 cols and 2654208 nnz values
Create sparse matrix map#
sparse_map = sp.sparse.coo_matrix((nnzvals, (rows, cols)), shape=(n_b, n_a))
sparse_map
<884736x777602 sparse matrix of type '<class 'numpy.float64'>'
with 2654208 stored elements in COOrdinate format>
Read in CAM-SE output#
tsfile = xr.open_dataset(data_path + data_file, chunks={"time": 50, "ncol": 50000})
tsfile
<xarray.Dataset>
Dimensions: (lev: 30, ilev: 31, ncol: 777602, time: 2920, nbnd: 2)
Coordinates:
* lev (lev) float64 3.643 7.595 14.36 24.61 ... 957.5 976.3 992.6
* ilev (ilev) float64 2.255 5.032 10.16 18.56 ... 967.5 985.1 1e+03
* time (time) object 2000-01-01 03:00:00 ... 2001-01-01 00:00:00
Dimensions without coordinates: ncol, nbnd
Data variables: (12/31)
hyam (lev) float64 dask.array<chunksize=(30,), meta=np.ndarray>
hybm (lev) float64 dask.array<chunksize=(30,), meta=np.ndarray>
P0 float64 ...
hyai (ilev) float64 dask.array<chunksize=(31,), meta=np.ndarray>
hybi (ilev) float64 dask.array<chunksize=(31,), meta=np.ndarray>
lat (ncol) float64 dask.array<chunksize=(50000,), meta=np.ndarray>
... ...
n2ovmr (time) float64 dask.array<chunksize=(50,), meta=np.ndarray>
f11vmr (time) float64 dask.array<chunksize=(50,), meta=np.ndarray>
f12vmr (time) float64 dask.array<chunksize=(50,), meta=np.ndarray>
sol_tsi (time) float64 dask.array<chunksize=(50,), meta=np.ndarray>
nsteph (time) int32 dask.array<chunksize=(50,), meta=np.ndarray>
TS (time, ncol) float32 dask.array<chunksize=(50, 50000), meta=np.ndarray>
Attributes:
np: 4
ne: 120
Conventions: CF-1.0
source: CAM
case: b.e13.B20TRC5CN.ne120_g16.003
title: UNSET
logname:
host:
Version: $Name$
revision_Id: $Id$
initial_file: /projects/ccsm/inputdata/atm/cam/inic/homme/cami-mam3_0...
topography_file: /projects/ccsm/inputdata/atm/cam/topo/USGS-gtopo30_ne12...Extract surface temperature variable (TS)#
ts = tsfile.TS
ts
<xarray.DataArray 'TS' (time: 2920, ncol: 777602)>
dask.array<open_dataset-bb5b27d0821608d3fc5037525bd2f985TS, shape=(2920, 777602), dtype=float32, chunksize=(50, 50000), chunktype=numpy.ndarray>
Coordinates:
* time (time) object 2000-01-01 03:00:00 ... 2001-01-01 00:00:00
Dimensions without coordinates: ncol
Attributes:
units: K
long_name: Surface temperature (radiative)Apply sparse map onto first timestep of TS#
Note: this is a time consuming step, so demonstrating only the first time index here
%%time
field_target = sparse_map @ ts.isel(time=0)
CPU times: user 2.53 s, sys: 116 ms, total: 2.64 s
Wall time: 33 s
field_target.shape
(884736,)
Reshape 1-D vector to destination grid#
data_out = np.reshape(field_target, dst_grid_dims, order='F')
print(f"Reshaped array now has dimensions {dst_grid_dims}")
Reshaped array now has dimensions [1152 768]
Reshape destination grid to build structured data array#
lat2d = np.reshape(dstlat.values, dst_grid_dims, order='F')
lon2d = np.reshape(dstlon.values, dst_grid_dims, order='F')
print(lat2d.shape, lon2d.shape)
(1152, 768) (1152, 768)
Build data array with dimension and coordinates#
ts_remap = xr.DataArray(
data=data_out,
dims=["lon", "lat"],
coords=dict(
lon=(["lon"], lon2d[:, 0]),
lat=(["lat"], lat2d[0, :]),
),
)
ts_remap
<xarray.DataArray (lon: 1152, lat: 768)>
array([[244.04267883, 243.94797035, 243.71354076, ..., 231.61040262,
231.07736837, 231.86198425],
[244.04267883, 243.94817018, 243.71394044, ..., 231.61271724,
231.07852571, 231.86198425],
[244.04267883, 243.94837698, 243.71435405, ..., 231.61500008,
231.07966716, 231.86198425],
...,
[244.04267883, 243.95208424, 243.72176851, ..., 231.57748313,
231.06090852, 231.86198425],
[244.04267883, 243.95070604, 243.71901211, ..., 231.58848746,
231.06641072, 231.86198425],
[244.04267883, 243.94933473, 243.7162695 , ..., 231.59946073,
231.07189739, 231.86198425]])
Coordinates:
* lon (lon) float64 0.0 0.3125 0.625 0.9375 ... 358.8 359.1 359.4 359.7
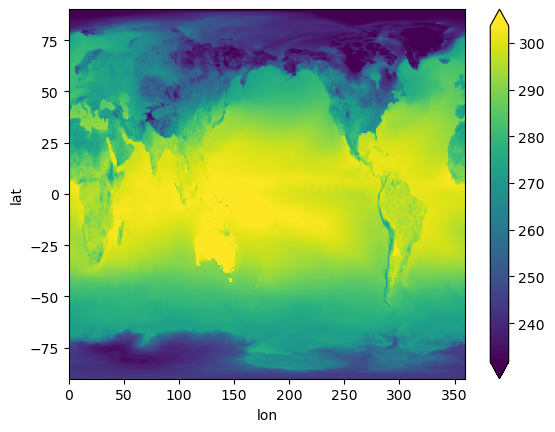
* lat (lat) float64 -90.0 -89.77 -89.53 -89.3 ... 89.3 89.53 89.77 90.0Visualize#
%%time
ts_remap.plot(x="lon", y="lat", robust=True);
CPU times: user 392 ms, sys: 47 ms, total: 439 ms
Wall time: 2.61 s
2. Use a CAM-SE remap function to scale up#
This method builds on the first example, by creating a function to streamline the remapping process. See related regridding script.
Define a function to remap using weights file#
def remap_camse(ds, dsw, varlst=[]):
# dso = xr.full_like(ds.drop_dims('ncol'), np.nan)
dso = ds.drop_dims('ncol').copy()
lonb = dsw.xc_b.values.reshape([dsw.dst_grid_dims[1].values, dsw.dst_grid_dims[0].values])
latb = dsw.yc_b.values.reshape([dsw.dst_grid_dims[1].values, dsw.dst_grid_dims[0].values])
weights = sp.sparse.coo_matrix(
(dsw.S, (dsw.row - 1, dsw.col - 1)), shape=[dsw.dims['n_b'], dsw.dims['n_a']]
)
if not varlst:
for varname in list(ds):
if 'ncol' in (ds[varname].dims):
varlst.append(varname)
if 'lon' in varlst:
varlst.remove('lon')
if 'lat' in varlst:
varlst.remove('lat')
if 'area' in varlst:
varlst.remove('area')
for varname in varlst:
shape = ds[varname].shape
invar_flat = ds[varname].values.reshape(-1, shape[-1])
remapped_flat = weights.dot(invar_flat.T).T
remapped = remapped_flat.reshape(
[*shape[0:-1], dsw.dst_grid_dims[1].values, dsw.dst_grid_dims[0].values]
)
dimlst = list(ds[varname].dims[0:-1])
dims = {}
coords = {}
for it in dimlst:
dims[it] = dso.dims[it]
coords[it] = dso.coords[it]
dims['lat'] = int(dsw.dst_grid_dims[1])
dims['lon'] = int(dsw.dst_grid_dims[0])
coords['lat'] = latb[:, 0]
coords['lon'] = lonb[0, :]
remapped = xr.DataArray(remapped, coords=coords, dims=dims, attrs=ds[varname].attrs)
dso = xr.merge([dso, remapped.to_dataset(name=varname)])
return dso
Read in CAM-SE output#
tsfile = xr.open_dataset(data_path + data_file, chunks={"time": 50})
tsfile
<xarray.Dataset>
Dimensions: (lev: 30, ilev: 31, ncol: 777602, time: 2920, nbnd: 2)
Coordinates:
* lev (lev) float64 3.643 7.595 14.36 24.61 ... 957.5 976.3 992.6
* ilev (ilev) float64 2.255 5.032 10.16 18.56 ... 967.5 985.1 1e+03
* time (time) object 2000-01-01 03:00:00 ... 2001-01-01 00:00:00
Dimensions without coordinates: ncol, nbnd
Data variables: (12/31)
hyam (lev) float64 dask.array<chunksize=(30,), meta=np.ndarray>
hybm (lev) float64 dask.array<chunksize=(30,), meta=np.ndarray>
P0 float64 ...
hyai (ilev) float64 dask.array<chunksize=(31,), meta=np.ndarray>
hybi (ilev) float64 dask.array<chunksize=(31,), meta=np.ndarray>
lat (ncol) float64 dask.array<chunksize=(777602,), meta=np.ndarray>
... ...
n2ovmr (time) float64 dask.array<chunksize=(50,), meta=np.ndarray>
f11vmr (time) float64 dask.array<chunksize=(50,), meta=np.ndarray>
f12vmr (time) float64 dask.array<chunksize=(50,), meta=np.ndarray>
sol_tsi (time) float64 dask.array<chunksize=(50,), meta=np.ndarray>
nsteph (time) int32 dask.array<chunksize=(50,), meta=np.ndarray>
TS (time, ncol) float32 dask.array<chunksize=(50, 777602), meta=np.ndarray>
Attributes:
np: 4
ne: 120
Conventions: CF-1.0
source: CAM
case: b.e13.B20TRC5CN.ne120_g16.003
title: UNSET
logname:
host:
Version: $Name$
revision_Id: $Id$
initial_file: /projects/ccsm/inputdata/atm/cam/inic/homme/cami-mam3_0...
topography_file: /projects/ccsm/inputdata/atm/cam/topo/USGS-gtopo30_ne12...Read in map file (weights)#
weights = xr.open_dataset(map_path + map_file)
weights
<xarray.Dataset>
Dimensions: (src_grid_rank: 1, dst_grid_rank: 2, n_a: 777602,
n_b: 884736, nv_a: 3, nv_b: 4, n_s: 2654208)
Dimensions without coordinates: src_grid_rank, dst_grid_rank, n_a, n_b, nv_a,
nv_b, n_s
Data variables: (12/19)
src_grid_dims (src_grid_rank) int32 ...
dst_grid_dims (dst_grid_rank) int32 ...
yc_a (n_a) float64 ...
yc_b (n_b) float64 ...
xc_a (n_a) float64 ...
xc_b (n_b) float64 ...
... ...
area_b (n_b) float64 ...
frac_a (n_a) float64 ...
frac_b (n_b) float64 ...
col (n_s) int32 ...
row (n_s) int32 ...
S (n_s) float64 ...
Attributes:
title: ESMF Offline Regridding Weight Generator
normalization: destarea
map_method: Bilinear remapping
ESMF_regrid_method: Bilinear
conventions: NCAR-CSM
domain_a: /glade/p/cgd/ccr/people/nanr/mapfiles/ne120.nc
domain_b: /glade/p/cgd/ccr/people/nanr/mapfiles/0.23x0.31.nc
grid_file_src: /glade/p/cgd/ccr/people/nanr/mapfiles/ne120.nc
grid_file_dst: /glade/p/cgd/ccr/people/nanr/mapfiles/0.23x0.31.nc
CVS_revision: 6.3.0rApply remap function onto first timestep of TS#
Note: this is a time consuming step, so demonstrating only the first time index here
%%time
out = remap_camse(tsfile.isel(time=0), weights) # single timestep
CPU times: user 56.2 ms, sys: 637 ms, total: 693 ms
Wall time: 890 ms
out
<xarray.Dataset>
Dimensions: (lev: 30, ilev: 31, nbnd: 2, lat: 768, lon: 1152)
Coordinates:
* lev (lev) float64 3.643 7.595 14.36 24.61 ... 957.5 976.3 992.6
* ilev (ilev) float64 2.255 5.032 10.16 18.56 ... 967.5 985.1 1e+03
time object 2000-01-01 03:00:00
* lat (lat) float64 -90.0 -89.77 -89.53 -89.3 ... 89.53 89.77 90.0
* lon (lon) float64 0.0 0.3125 0.625 0.9375 ... 359.1 359.4 359.7
Dimensions without coordinates: nbnd
Data variables: (12/28)
hyam (lev) float64 dask.array<chunksize=(30,), meta=np.ndarray>
hybm (lev) float64 dask.array<chunksize=(30,), meta=np.ndarray>
P0 float64 ...
hyai (ilev) float64 dask.array<chunksize=(31,), meta=np.ndarray>
hybi (ilev) float64 dask.array<chunksize=(31,), meta=np.ndarray>
ntrm int32 ...
... ...
n2ovmr float64 dask.array<chunksize=(), meta=np.ndarray>
f11vmr float64 dask.array<chunksize=(), meta=np.ndarray>
f12vmr float64 dask.array<chunksize=(), meta=np.ndarray>
sol_tsi float64 dask.array<chunksize=(), meta=np.ndarray>
nsteph int32 dask.array<chunksize=(), meta=np.ndarray>
TS (lat, lon) float64 244.0 244.0 244.0 ... 231.9 231.9 231.9
Attributes:
np: 4
ne: 120
Conventions: CF-1.0
source: CAM
case: b.e13.B20TRC5CN.ne120_g16.003
title: UNSET
logname:
host:
Version: $Name$
revision_Id: $Id$
initial_file: /projects/ccsm/inputdata/atm/cam/inic/homme/cami-mam3_0...
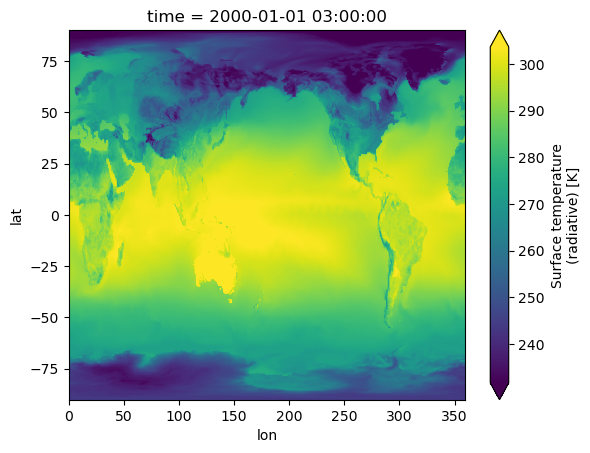
topography_file: /projects/ccsm/inputdata/atm/cam/topo/USGS-gtopo30_ne12...Visualize#
%%time
out.TS.plot(robust=True);
CPU times: user 112 ms, sys: 1.47 ms, total: 114 ms
Wall time: 115 ms
3. Regrid CAM-SE output using xESMF#
This method utilizes the xESMF package which has built-in regridding functionality. See a related discussion and example in this ESDS blog post.
Define regridding function that constructs an xESMF regridder#
def regrid_cam_se(dataset, weight_file):
"""
Regrid CAM-SE output using an existing ESMF weights file.
Parameters
----------
dataset: xarray.Dataset
Input dataset to be regridded. Must have the `ncol` dimension.
weight_file: str or Path
Path to existing ESMF weights file
Returns
-------
regridded
xarray.Dataset after regridding.
"""
import numpy as np
import xarray as xr
assert isinstance(dataset, xr.Dataset)
weights = xr.open_dataset(weight_file)
# input variable shape
in_shape = weights.src_grid_dims.load().data
# Since xESMF expects 2D vars, we'll insert a dummy dimension of size-1
if len(in_shape) == 1:
in_shape = [1, in_shape.item()]
# output variable shapew
out_shape = weights.dst_grid_dims.load().data.tolist()[::-1]
print(f"Regridding from {in_shape} to {out_shape}")
# Insert dummy dimension
vars_with_ncol = [name for name in dataset.variables if "ncol" in dataset[name].dims]
updated = dataset.copy().update(
dataset[vars_with_ncol].transpose(..., "ncol").expand_dims("dummy", axis=-2)
)
# construct a regridder
# use empty variables to tell xesmf the right shape
# https://github.com/pangeo-data/xESMF/issues/202
dummy_in = xr.Dataset(
{
"lat": ("lat", np.empty((in_shape[0],))),
"lon": ("lon", np.empty((in_shape[1],))),
}
)
dummy_out = xr.Dataset(
{
"lat": ("lat", weights.yc_b.data.reshape(out_shape)[:, 0]),
"lon": ("lon", weights.xc_b.data.reshape(out_shape)[0, :]),
}
)
regridder = xesmf.Regridder(
dummy_in,
dummy_out,
weights=weight_file,
method="test",
reuse_weights=True,
periodic=True,
)
display(regridder)
# Actually regrid, after renaming
regridded = regridder(updated.rename({"dummy": "lat", "ncol": "lon"}))
# merge back any variables that didn't have the ncol dimension
# And so were not regridded
return xr.merge([dataset.drop_vars(regridded.variables), regridded])
Read in CAM-SE output#
tsfile = xr.open_dataset(data_path + data_file, chunks={"time": 50, "ncol": -1})
Apply the regridder#
regridded = regrid_cam_se(tsfile, f"{map_path}/{map_file}")
regridded
Regridding from [1, 777602] to [768, 1152]
xESMF Regridder
Regridding algorithm: test
Weight filename: test_1x777602_768x1152_peri.nc
Reuse pre-computed weights? True
Input grid shape: (1, 777602)
Output grid shape: (768, 1152)
Periodic in longitude? True
<xarray.Dataset>
Dimensions: (lev: 30, ilev: 31, time: 2920, nbnd: 2, lat: 768, lon: 1152)
Coordinates:
* lev (lev) float64 3.643 7.595 14.36 24.61 ... 957.5 976.3 992.6
* ilev (ilev) float64 2.255 5.032 10.16 18.56 ... 967.5 985.1 1e+03
* time (time) object 2000-01-01 03:00:00 ... 2001-01-01 00:00:00
* lat (lat) float64 -90.0 -89.77 -89.53 -89.3 ... 89.53 89.77 90.0
* lon (lon) float64 0.0 0.3125 0.625 0.9375 ... 359.1 359.4 359.7
Dimensions without coordinates: nbnd
Data variables: (12/29)
hyam (lev) float64 dask.array<chunksize=(30,), meta=np.ndarray>
hybm (lev) float64 dask.array<chunksize=(30,), meta=np.ndarray>
P0 float64 ...
hyai (ilev) float64 dask.array<chunksize=(31,), meta=np.ndarray>
hybi (ilev) float64 dask.array<chunksize=(31,), meta=np.ndarray>
ntrm int32 ...
... ...
f11vmr (time) float64 dask.array<chunksize=(50,), meta=np.ndarray>
f12vmr (time) float64 dask.array<chunksize=(50,), meta=np.ndarray>
sol_tsi (time) float64 dask.array<chunksize=(50,), meta=np.ndarray>
nsteph (time) int32 dask.array<chunksize=(50,), meta=np.ndarray>
area (lat, lon) float64 dask.array<chunksize=(768, 1152), meta=np.ndarray>
TS (time, lat, lon) float32 dask.array<chunksize=(50, 768, 1152), meta=np.ndarray>
Attributes:
np: 4
ne: 120
Conventions: CF-1.0
source: CAM
case: b.e13.B20TRC5CN.ne120_g16.003
title: UNSET
logname:
host:
Version: $Name$
revision_Id: $Id$
initial_file: /projects/ccsm/inputdata/atm/cam/inic/homme/cami-mam3_0...
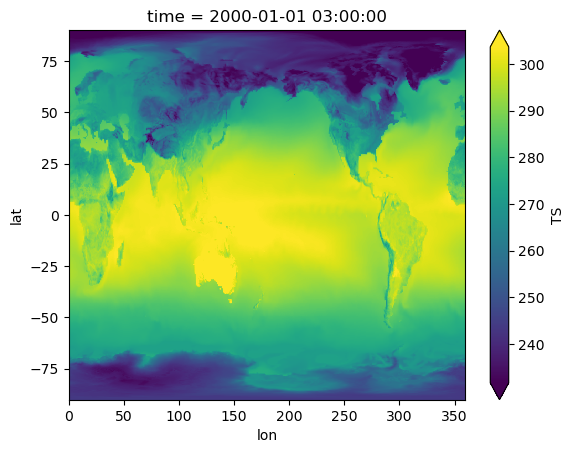
topography_file: /projects/ccsm/inputdata/atm/cam/topo/USGS-gtopo30_ne12...Visualize#
%%time
regridded.TS.isel(time=0).plot(robust=True);
CPU times: user 528 ms, sys: 95 ms, total: 623 ms
Wall time: 10.5 s
4. Direct sparse matrix multiply-add#
This method uses the weights to create a pydata/sparse matrix, then uses xr.dot (≡ np.einsum). This approach is identical to methods 1, 2; but with pydata/sparse which in theory should work well with Dask. This should support chunking along ncol but doesn’t work because of a sparse bug. So instead we work-around the bug using the very cool opt_einsum package.
Define a function that reads in map file (weights) and constructs a sparse array#
def read_xesmf_weights_file(filename):
import numpy as np
import sparse
import xarray as xr
weights = xr.open_dataset(filename)
# input variable shape
in_shape = weights.src_grid_dims.load().data
# output variable shape
out_shape = weights.dst_grid_dims.load().data.tolist()[::-1]
print(f"Regridding from {in_shape} to {out_shape}")
rows = weights['row'] - 1 # row indices (1-based)
cols = weights['col'] - 1 # col indices (1-based)
# construct a sparse array,
# reshape to 3D : lat, lon, ncol
# This reshaping should allow optional chunking along
# lat, lon later
sparse_array_data = sparse.COO(
coords=np.stack([rows.data, cols.data]),
data=weights.S.data,
shape=(weights.sizes["n_b"], weights.sizes["n_a"]),
fill_value=0,
).reshape((*out_shape, -1))
# Create a DataArray with sparse weights and the output coordinates
xsparse_wgts = xr.DataArray(
sparse_array_data,
dims=("lat", "lon", "ncol"),
# Add useful coordinate information, this will get propagated to the output
coords={
"lat": ("lat", weights.yc_b.data.reshape(out_shape)[:, 0]),
"lon": ("lon", weights.xc_b.data.reshape(out_shape)[0, :]),
},
# propagate useful information like regridding algorithm
attrs=weights.attrs,
)
return xsparse_wgts
Read in weights#
xsparse_wgts = read_xesmf_weights_file(map_path + map_file)
xsparse_wgts
Regridding from [777602] to [768, 1152]
<xarray.DataArray (lat: 768, lon: 1152, ncol: 777602)>
<COO: shape=(768, 1152, 777602), dtype=float64, nnz=2654208, fill_value=0.0>
Coordinates:
* lat (lat) float64 -90.0 -89.77 -89.53 -89.3 ... 89.3 89.53 89.77 90.0
* lon (lon) float64 0.0 0.3125 0.625 0.9375 ... 358.8 359.1 359.4 359.7
Dimensions without coordinates: ncol
Attributes:
title: ESMF Offline Regridding Weight Generator
normalization: destarea
map_method: Bilinear remapping
ESMF_regrid_method: Bilinear
conventions: NCAR-CSM
domain_a: /glade/p/cgd/ccr/people/nanr/mapfiles/ne120.nc
domain_b: /glade/p/cgd/ccr/people/nanr/mapfiles/0.23x0.31.nc
grid_file_src: /glade/p/cgd/ccr/people/nanr/mapfiles/ne120.nc
grid_file_dst: /glade/p/cgd/ccr/people/nanr/mapfiles/0.23x0.31.nc
CVS_revision: 6.3.0rRead in CAM-SE output#
tsfile = xr.open_dataset(data_path + data_file, chunks={"time": 1, "ncol": -1})
tsfile
<xarray.Dataset>
Dimensions: (lev: 30, ilev: 31, ncol: 777602, time: 2920, nbnd: 2)
Coordinates:
* lev (lev) float64 3.643 7.595 14.36 24.61 ... 957.5 976.3 992.6
* ilev (ilev) float64 2.255 5.032 10.16 18.56 ... 967.5 985.1 1e+03
* time (time) object 2000-01-01 03:00:00 ... 2001-01-01 00:00:00
Dimensions without coordinates: ncol, nbnd
Data variables: (12/31)
hyam (lev) float64 dask.array<chunksize=(30,), meta=np.ndarray>
hybm (lev) float64 dask.array<chunksize=(30,), meta=np.ndarray>
P0 float64 ...
hyai (ilev) float64 dask.array<chunksize=(31,), meta=np.ndarray>
hybi (ilev) float64 dask.array<chunksize=(31,), meta=np.ndarray>
lat (ncol) float64 dask.array<chunksize=(777602,), meta=np.ndarray>
... ...
n2ovmr (time) float64 dask.array<chunksize=(1,), meta=np.ndarray>
f11vmr (time) float64 dask.array<chunksize=(1,), meta=np.ndarray>
f12vmr (time) float64 dask.array<chunksize=(1,), meta=np.ndarray>
sol_tsi (time) float64 dask.array<chunksize=(1,), meta=np.ndarray>
nsteph (time) int32 dask.array<chunksize=(1,), meta=np.ndarray>
TS (time, ncol) float32 dask.array<chunksize=(1, 777602), meta=np.ndarray>
Attributes:
np: 4
ne: 120
Conventions: CF-1.0
source: CAM
case: b.e13.B20TRC5CN.ne120_g16.003
title: UNSET
logname:
host:
Version: $Name$
revision_Id: $Id$
initial_file: /projects/ccsm/inputdata/atm/cam/inic/homme/cami-mam3_0...
topography_file: /projects/ccsm/inputdata/atm/cam/topo/USGS-gtopo30_ne12...Compute#
Now that we have weights, we compute the “dot product” using xr.dot. xr.dot uses numpy.einsum under the hood, which dispatches to sparse.einsum. If we do this we get an error due to a bug.
%xmode Minimal
regridded = xr.dot(
tsfile.TS,
# could chunk differently here
xsparse_wgts.chunk(),
# This dimension will be "contracted"
# or summmed over after multiplying by the weights
dims="ncol",
)
regridded
Exception reporting mode: Minimal
TypeError: einsum() got an unexpected keyword argument 'dtype'
Instead use opt_einsum#
Turns out we can use the very cool opt_einsum package instead. opt_einsum.contract is a drop-in replacement for numpy.einsum (which powers xr.dot) so we’ll just monkey-patch this in to avoid redoing a bunch of xarray code that converts dimension names to the “subscript string” that opt_einsum.contract expects. Seems like xarray could support opt_einsum as an opt-in.
opt_einsum.contract("tc,yxc->tyx", tsfile.TS.data, xsparse_wgts.chunk().data)
|
||||||||||||||||
Define a function to apply weights using this method#
def apply_weights(dataset, weights):
def _apply(da):
# 🐵 🔧
xr.core.duck_array_ops.einsum = opt_einsum.contract
ans = xr.dot(
da,
weights,
# This dimension will be "contracted"
# or summmed over after multiplying by the weights
dims="ncol",
)
# 🐵 🔧 : restore back to original
xr.core.duck_array_ops.einsum = np.einsum
return ans
vars_with_ncol = [
name
for name, array in dataset.variables.items()
if "ncol" in array.dims and name not in weights.coords
]
regridded = dataset[vars_with_ncol].map(_apply)
# merge in other variables, but skip those that are already set
# like lat, lon
return xr.merge([dataset.drop_vars(regridded.variables), regridded])
Apply weights#
regridded = apply_weights(tsfile, xsparse_wgts.chunk())
regridded
<xarray.Dataset>
Dimensions: (lev: 30, ilev: 31, time: 2920, nbnd: 2, lat: 768, lon: 1152)
Coordinates:
* lev (lev) float64 3.643 7.595 14.36 24.61 ... 957.5 976.3 992.6
* ilev (ilev) float64 2.255 5.032 10.16 18.56 ... 967.5 985.1 1e+03
* time (time) object 2000-01-01 03:00:00 ... 2001-01-01 00:00:00
* lat (lat) float64 -90.0 -89.77 -89.53 -89.3 ... 89.53 89.77 90.0
* lon (lon) float64 0.0 0.3125 0.625 0.9375 ... 359.1 359.4 359.7
Dimensions without coordinates: nbnd
Data variables: (12/29)
hyam (lev) float64 dask.array<chunksize=(30,), meta=np.ndarray>
hybm (lev) float64 dask.array<chunksize=(30,), meta=np.ndarray>
P0 float64 ...
hyai (ilev) float64 dask.array<chunksize=(31,), meta=np.ndarray>
hybi (ilev) float64 dask.array<chunksize=(31,), meta=np.ndarray>
ntrm int32 ...
... ...
f11vmr (time) float64 dask.array<chunksize=(1,), meta=np.ndarray>
f12vmr (time) float64 dask.array<chunksize=(1,), meta=np.ndarray>
sol_tsi (time) float64 dask.array<chunksize=(1,), meta=np.ndarray>
nsteph (time) int32 dask.array<chunksize=(1,), meta=np.ndarray>
area (lat, lon) float64 dask.array<chunksize=(768, 1152), meta=np.ndarray>
TS (time, lat, lon) float64 dask.array<chunksize=(1, 768, 1152), meta=np.ndarray>
Attributes:
np: 4
ne: 120
Conventions: CF-1.0
source: CAM
case: b.e13.B20TRC5CN.ne120_g16.003
title: UNSET
logname:
host:
Version: $Name$
revision_Id: $Id$
initial_file: /projects/ccsm/inputdata/atm/cam/inic/homme/cami-mam3_0...
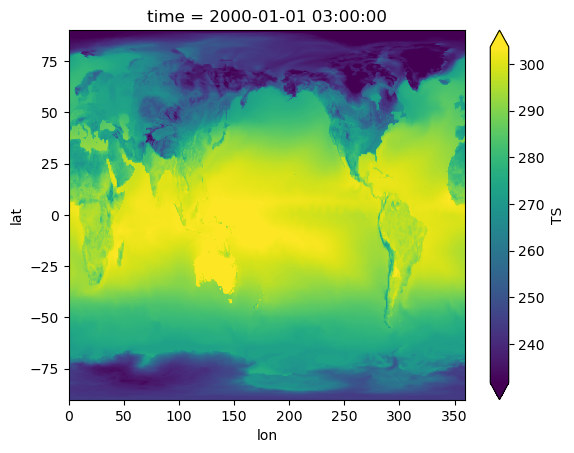
topography_file: /projects/ccsm/inputdata/atm/cam/topo/USGS-gtopo30_ne12...Visualize#
%%time
regridded.TS.isel(time=0).compute().plot(robust=True);
CPU times: user 223 ms, sys: 74.8 ms, total: 298 ms
Wall time: 878 ms
Chunking in space#
Let’s try chunking along lon. This is useless since you’ll always read all ncol in, but it illustrates the point. Note that these chunk sizes are very small, so this is probably only useful for a much higher-resolution output grid.
regridded = apply_weights(tsfile, xsparse_wgts.chunk(lon=200))
regridded
<xarray.Dataset>
Dimensions: (lev: 30, ilev: 31, time: 2920, nbnd: 2, lat: 768, lon: 1152)
Coordinates:
* lev (lev) float64 3.643 7.595 14.36 24.61 ... 957.5 976.3 992.6
* ilev (ilev) float64 2.255 5.032 10.16 18.56 ... 967.5 985.1 1e+03
* time (time) object 2000-01-01 03:00:00 ... 2001-01-01 00:00:00
* lat (lat) float64 -90.0 -89.77 -89.53 -89.3 ... 89.53 89.77 90.0
* lon (lon) float64 0.0 0.3125 0.625 0.9375 ... 359.1 359.4 359.7
Dimensions without coordinates: nbnd
Data variables: (12/29)
hyam (lev) float64 dask.array<chunksize=(30,), meta=np.ndarray>
hybm (lev) float64 dask.array<chunksize=(30,), meta=np.ndarray>
P0 float64 ...
hyai (ilev) float64 dask.array<chunksize=(31,), meta=np.ndarray>
hybi (ilev) float64 dask.array<chunksize=(31,), meta=np.ndarray>
ntrm int32 ...
... ...
f11vmr (time) float64 dask.array<chunksize=(1,), meta=np.ndarray>
f12vmr (time) float64 dask.array<chunksize=(1,), meta=np.ndarray>
sol_tsi (time) float64 dask.array<chunksize=(1,), meta=np.ndarray>
nsteph (time) int32 dask.array<chunksize=(1,), meta=np.ndarray>
area (lat, lon) float64 dask.array<chunksize=(768, 200), meta=np.ndarray>
TS (time, lat, lon) float64 dask.array<chunksize=(1, 768, 200), meta=np.ndarray>
Attributes:
np: 4
ne: 120
Conventions: CF-1.0
source: CAM
case: b.e13.B20TRC5CN.ne120_g16.003
title: UNSET
logname:
host:
Version: $Name$
revision_Id: $Id$
initial_file: /projects/ccsm/inputdata/atm/cam/inic/homme/cami-mam3_0...
topography_file: /projects/ccsm/inputdata/atm/cam/topo/USGS-gtopo30_ne12...Visualize#
%%time
regridded.TS.isel(time=0).compute().plot(robust=True);
CPU times: user 582 ms, sys: 96.9 ms, total: 679 ms
Wall time: 3.67 s

Timing performance for core multiply-add#
Testing the model or the core operation that would happen on every dask block. data.data is a numpy array, and xsparse_weights.data is a sparse.COO array.
Does the order of operations matter?
data = tsfile.TS.isel(time=[0]).load()
%timeit np.einsum('tc, yxc->tyx', data.data, xsparse_wgts.data).todense()
%timeit opt_einsum.contract('tc, yxc->tyx', data.data, xsparse_wgts.data)
13.7 s ± 748 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
82.5 ms ± 789 µs per loop (mean ± std. dev. of 7 runs, 1 loop each)
%timeit np.einsum('yxc, tc->yxt', xsparse_wgts.data, data.data).todense()
%timeit opt_einsum.contract('yxc, tc->yxt', xsparse_wgts.data, data.data)
146 ms ± 3.02 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)
279 ms ± 7.15 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
Summary of the timing test:#
Order of operations matters, particularly when calling
np.einsum.For best performance use
opt_einsum.contractand provide the field variable first, and weights second.
Summary#
In this blog post we’ve explored 4 methods to remap and visualize CAM-SE unstructured output in Python. Methods 1 and 2 provide a basic version of remapping using an ESMF weights file, mimicking the NCO tool ncremap while allowing the user to perform the remapping in a Python or Jupyter notebook environment. Method 3 demonstrates the use of xESMF for this purpose, with similar performance. Method 4 expands on the approach of methods 1 and 2 to allow for scaling with Dask.
We look forward to feedback from users on this code, and in particular which methods work well for different applications!
Further reading#
Here are some additional methods to explore that came up in discussions around this topic. Let us know if you try these out!