2: Use the BGC model#
We can use a different I compset: IHistClm50BgcCrop. This experiment is a 20th century transient run using GSWP3v1 and the biogeochemistry model including crops.
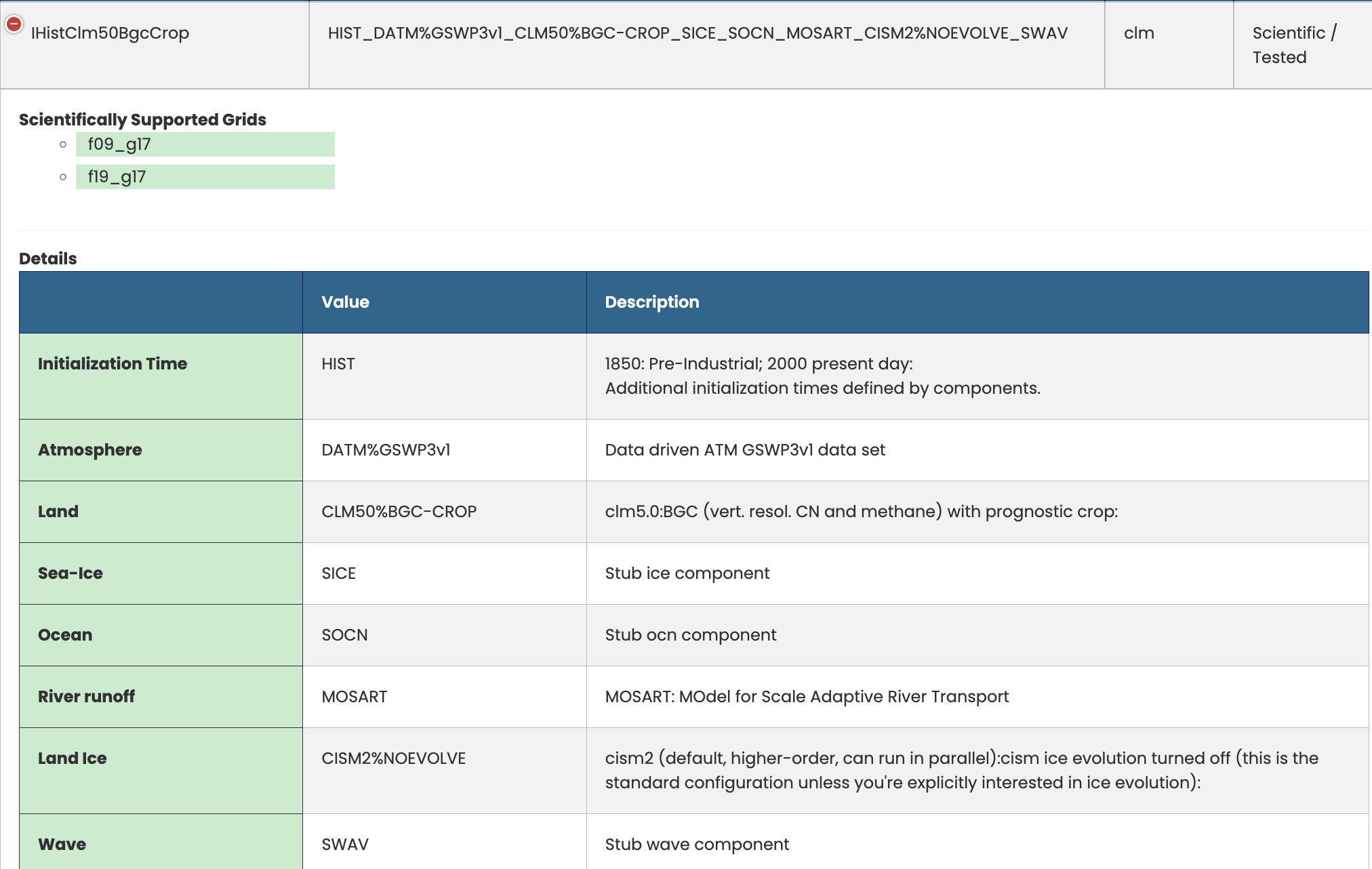
Figure: IHIST compset definition.
Create a case called i.day5.b using the compset IHistClm50BgcCrop at f09_g17_gl4 resolution.
Set the run length to 5 days.
Build and run the model.
Click here for the solution
Create a new case i.day5.b :
cd /glade/u/home/$USER/code/my_cesm_code/cime/scripts
./create_newcase --case ~/cases/i.day5.b --compset IHistClm50BgcCrop --res f09_g17_gl4 --run-unsupported
Case setup:
cd ~/cases/i.day5.b
./case.setup
Note differences between this case and the control case:
diff env_run.xml ../i.day5.a/env_run.xml
Change the clm namelist using user_nl_clm by adding the following lines:
hist_nhtfrq = -24
hist_mfilt = 6
Check the namelist by running:
./preview_namelists
If needed, change job queue, account number, or wallclock time. For instance:
./xmlchange JOB_QUEUE=tutorial,PROJECT=UESM0014 --force,JOB_WALLCLOCK_TIME=0:15:00
Build case:
qcmd -- ./case.build
Compare the namelists from the two experiments:
diff CaseDocs/lnd_in ../i.day5.a/CaseDocs/lnd_in
Submit case:
./case.submit
When the run is completed, look into the archive directory for: i.day5.b.
(1) Check that your archive directory on derecho (The path will be different on other machines):
cd /glade/derecho/scratch/$USER/archive/i.day5.b/lnd/hist
ls
(2) Compare to control run:
ncdiff -v TLAI i.day5.b.clm2.XXX.nc /glade/derecho/scratch/$USER/archive/i.day5.a/lnd/hist/i.day5.a.clm2.XXX.nc i_diff.nc
ncview i_diff.nc
Test your understanding#
What changes do you see from the control case with the prognostic BGC?
… OTHERS?
